- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2000
- Life Sciences
- Structure of DNA Mismatch Repair Protein MutS Binding to a G:T Mismatch
Structure of DNA Mismatch Repair Protein MutS Binding to a G:T Mismatch
DNA mismatch repair (MMR) is responsible for the maintenance of DNA fidelity upon replication. It captures errors in the newly synthesised DNA strand that are missed by the polymerase proofreading, lowering the mutation frequency by a factor of 100-1000. In humans, loss of a single allele of one of the mismatch repair proteins causes one of the most widespread forms of predisposition for cancer, called hereditary non-polyposis colon cancer (HNPCC). Functional mismatch repair is dependent on recognition of mismatches by a dimeric MutS protein, a homodimer in bacteria but a heterodimer in humans (MSH2/MSH6 or MSH2/MSH3). Upon recognition of the mismatch and binding of ATP, MutS can activate a second protein MutL to search for a strand discrimination signal. Once this signal is found (up to 1 kb away) the newly synthesised strand is removed and resynthesised.
The crystal structure of Escherichia coli MutS has now been solved. Crystals were grown of C800, a C-terminally truncated (53 amino acids) MutS, in the presence of a G:T mismatched DNA oligomer. The structure was solved with a three-wavelength selenomethionine MAD experiment, performed at beamline BM14. High-resolution data (2.2 Å) from the same crystal were collected at beam line ID14-2.
In the structure (Figure 2) dimeric MutS is bound to one mismatched DNA oligomer. The protein embraces the DNA with non-specific binding domains from both monomers, while it probes the mismatch with the N-terminal domain from just one monomer. Thus this protein exists as a structural heterodimer, explaining why only one of the partners in the eukaryotic heterodimers determines which mismatch is bound. The mismatch is recognised by hydrogen bonds to the bases and insertion of a phenylalanine, which sharply kinks the DNA at the mismatch site. The flexible DNA recognition domains allow MutS to accommodate the small changes in binding that are necessary to recognise all the different mismatches.
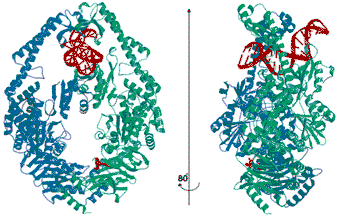 |
Fig. 2: Crystal structure of E. coli MutS. The MutS monomer (green) recognises the G:T mismatch in the DNA (red) and has ADP bound (at the bottom of the figure in red). The monomer in blue provides the non-specific DNA binding domains and an empty ATPase domain.
|
MutS has composite ATPase domains in which both monomers participate, with similarity to the ABC transporter ATPase domains. In the crystal structure only one monomer has ADP bound, indicating the existence of a complicated ATPase cycle, in which alternating ATPase activities in the two monomers play a role. This is similar to other multimeric ATPases, but had not been realised for the MutS family.
These structures provide a basis for understanding the precise role of MutS in DNA mismatch repair and help in the analysis the effect of specific mutations in HNPCC patients.
Principal Publication and Authors
M.H. Lamers (a), A. Perrakis (b), J.H. Enzlin (a), H.H.K. Winterwerp (a), N. de Wind (a), and T.K. Sixma (a), Nature, 407, 711-717 (2000).
(a) Netherlands Cancer Institute, Amsterdam
(The Netherlands)
(b) EMBL, Grenoble (France)



