- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2017
- Structural biology
- Structure and ceramidase function of adiponectin receptors
Structure and ceramidase function of adiponectin receptors
Crystal structures of adiponectin receptors (ADIPORs) are advancing understanding of the molecular mechanisms underlying their potent antidiabetic character. Here, structural data together with biochemical analyses reveal an intrinsic ceramidase activity of ADIPORs. This knowledge will facilitate structure-based drug design to treat obesity-related disease such as type 2 diabetes.
Adiponectin receptors (ADIPORs) are integral membrane proteins possessing a potent antidiabetic character, at least partly arbitrated by receptor-associated ceramidase activity, resulting in hydrolysis of ceramide to sphingosine and fatty acid.
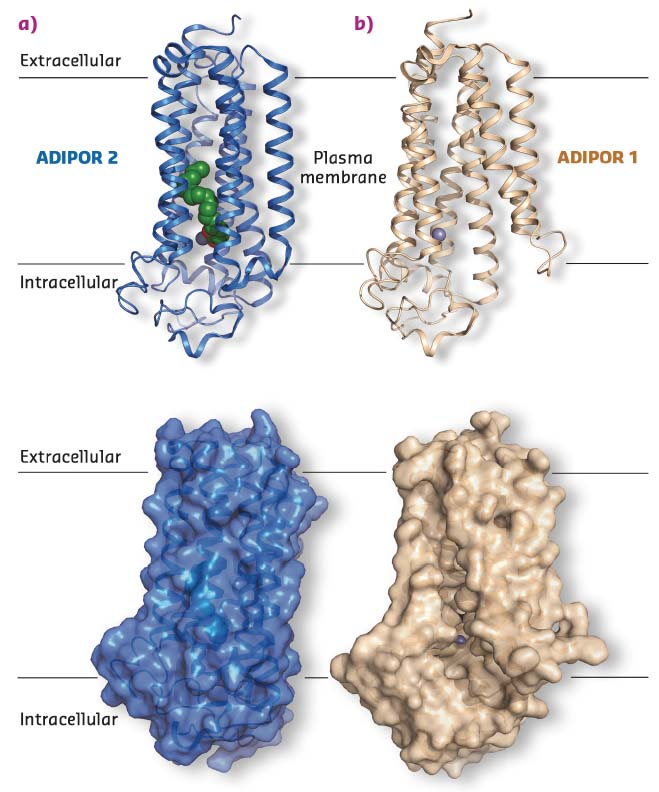
The recent crystal structures of two adiponectin receptor subtypes, ADIPOR1 and ADIPOR2, revealed that they possess seven transmembrane (7TM) helices enclosing large unoccupied internal cavities and a zinc binding site [1]. However, the molecular mechanisms of ADIPOR function were still obscure. In an effort to better understand how ADIPORs function, in meso crystallography was used and, following data collection at beamline ID30B, the structure of ADIPOR2 was determined (Figure 28a).
 |
|
Fig. 28: a) Crystal structure of ADIPOR2, blue cartoon (top) and surface view (bottom), with free fatty acid (green spheres) and a Zn2+ (grey sphere) binding site. b) Crystal structure of ADIPOR1, wheat cartoon (top) and surface view (bottom), with no free fatty acid bound and the Zn2+ (grey sphere) binding site exposed to the membrane environment. |
Besides the 7TM architecture and the position of the zinc binding site, the new ADIPOR2 crystal structures revealed the presence of a free fatty acid (FFA) within a large internal cavity. The FFA is bound close to a putative zinc catalytic core, prompting a test of whether ADIPORs possess an intrinsic ceramidase activity. To do so, purified receptor preparations were used to determine their ability to bind and hydrolyse synthetic ceramide substrates using biochemical approaches. In addition, computational studies provided key mechanistic insights into the ceramidase function of ADIPOR2, highlighting the central role played by the zinc catalytic core and surrounding amino acids.
Structural analysis naturally led to comparisons of data with previously published work. Surprisingly, it was observed that the electron density from a recently determined ADIPOR1 crystal structure appeared to have been misinterpreted, leading to a substantial error in the structural model produced [1]. Using the available structure factors this crystal structure was refined using the PDB entry 3WXV as a starting point, producing a final model with much improved statistics and fit to the electron density. In stark contrast to what had been published, the corrected ADIPOR1 structure is quite distinct from the ADIPOR2 structures obtained (Figure 28). Specifically, no FFA molecule is bound in the ADIPOR1 structure and both the ceramide binding pocket and the putative zinc catalytic site are wide open and accessible to the inner membrane leaflet (Figure 28b). Because both ADIPOR1 and ADIPOR2 possess an intrinsic ceramidase activity, it is suspected that the two structures may represent key steps in this process.
In conclusion, it has been demonstrated that ADIPORs possess an intrinsic ceramidase activity (i.e. hydrolyse ceramide into a free fatty acid (FFA) and sphingosine) and a combination of lipidic cubic phase crystallography and computational and biochemical studies provide the molecular basis for this enzymatic activity. Since insulin resistance and type 2 diabetes correlate with increased levels of ceramides and reduced expression of ADIPORs, this study may facilitate the development of new therapeutic approaches to treat obesity-related diseases.
Principal publication and authors
Structural insights into adiponectin receptors suggest ceramidase activity, I. Vasiliauskaité-Brooks (a), R. Sounier (a), P. Rochaix (a), G. Bellot (a), M. Fortier (a), F. Hoh (b), L. De Colibus (c), C. Bechara (d), E.M. Saied (e, f), C. Arenz (e), C. Leyrat (a) and S. Granier (a), Nature 544, 120-123 (2017); doi:10.1038/nature21714.
(a) Institut de Génomique Fonctionnelle, CNRS UMR-5203 INSERM U1191, University of Montpellier (France)
(b) Centre de Biochimie Structurale, CNRS UMR 5048-INSERM 1054 University of Montpellier (France)
(c) Division of Structural Biology, University of Oxford (UK)
(d) Dynamique des Interactions Membranaires Normales et Pathologiques, CNRS UMR5235, University of Montpellier (France)
(e) Institute for Chemistry, Humboldt-Universität zu Berlin (Germany)
(f) Chemistry Department, Faculty of Science, Suez Canal University, Ismailia (Egypt)
References
[1] H. Tanabe et al., Nature 520, 312-316 (2015).



