- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2015
- Matter at extremes
- Nuclear resonance scattering clarifies the structure of a pharmacologically relevant iron sulfur protein and its interaction with inhibitors
Nuclear resonance scattering clarifies the structure of a pharmacologically relevant iron sulfur protein and its interaction with inhibitors
The LytB/IspH protein catalyses the last step of the biochemical MEP-pathway used by most pathogenic bacteria for the biosynthesis of essential terpenoids. Due to its absence in humans, the MEP pathway is a target for the development of new antimicrobial agents. Nuclear resonance vibrational spectroscopy (NRVS) combined with quantum chemical-molecular mechanical (QM/MM) calculations have provided more insight into the active site structures of LytB in its substrate-free and inhibitor bound forms.
The LytB protein, also called IspH, catalyses the last step of the methylerythritol phosphate (MEP) pathway that carries out the biosynthesis of essential terpenoids in most pathogenic bacteria. The MEP pathway is essential for the survival of microorganisms, but it does not exist in mammals and humans. Humans too have to synthesise terpenoids such as cholesterol and hormones, but for this purpose a totally different biochemical pathway exists. Therefore, the MEP pathway is a promising target for the development of new antimicrobial agents. Inhibitors of the LytB protein may qualify for new potent antibiotics which are effective also for pathogens resistant to present drug therapies.
The LytB protein has a 4Fe-4S cluster which is essential for the catalytic activity of this enzyme, the conversion of (E)-4-hydroxy-3-methylbut-2-en-1-yl diphosphate (HMBPP, 1) into a mixture of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). The structure of the LytB protein has been solved but the reported structure for substrate free LytB shows a 3Fe-4S cluster.
In collaboration with the nuclear resonance scattering groups at beamlines ID18, ESRF, and P01, PETRA III, the structure of 4Fe-4S cluster in the substrate free LytB could be clarified. This was possible by sensing specifically the iron ligand vibrations by means of nuclear resonance vibrational spectroscopy (NRVS). NRVS, also called nuclear inelastic scattering (NIS) of synchrotron radiation, can be regarded as an extension of the conventional energy-resolved Mössbauer spectroscopy to energies in the order of molecular vibrations. The energy of the incoming synchrotron radiation is varied by high resolution monochromators within an interval of up to ±100 meV (± ~800 cm–1) or more around the resonance energy Eγ =14.4 keV of the 57Fe-Mössbauer nucleus with an experimental resolution < 1 meV (8 cm–1).
![The [4Fe-4S] cluster of the LytB/IspH protein](/files/live/sites/www/files/UsersAndScience/Publications/Highlights/2015/figure-108-HL-2015-opt.jpg) |
|
Fig. 108: Left: The [4Fe-4S] cluster of the LytB/IspH protein is essential for its enzymatic function. Its apical iron atom is linked to three water molecules. Right: This has been determined by NRVS experiments and corresponding simulations. |
Our experimental results, in combination with quantum chemical-molecular mechanical calculations on the whole protein, have shown that the substrate-free LytB protein contains an unusual [4Fe-4S]2+ cluster, the apical iron atom of which is linked to three water molecules (Figure 108). The presence of three labile water ligands on the apical iron may explain the instability of the [4Fe-4S]2+ cluster in crystallisation experiments causing the very high oxygen sensitivity of the isolated enzyme. To our knowledge, the coordination of the unique site of a 4Fe-4S cluster by three water molecules, as seen for LytB, is unprecedented. Whether the water ligands can also serve as proton donors for the enzymatic reaction of LytB is currently under investigation.
 |
|
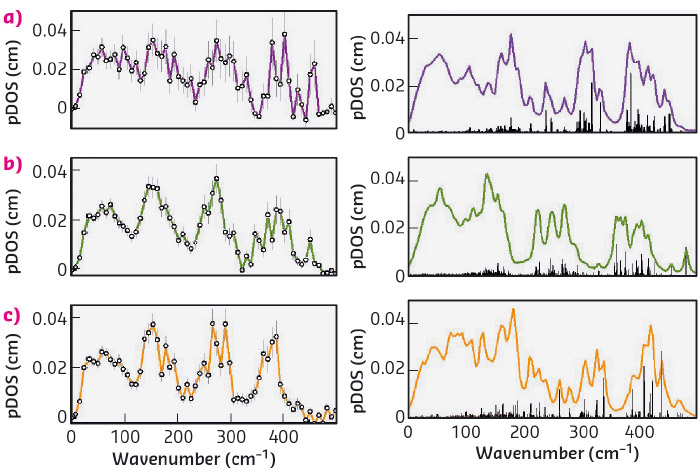
Fig. 109: Left: Partial density of states (pDOS) of the substrate-bound form of LytB (a), LytB in complex with inhibitor 2 (b), and LytB in complex with inhibitor 3 (c). Right: pDOS simulations assuming model structures of the active site/inhibitor complexes: (a) substrate-bound LytB; (b) amino inhibitor bound LytB (3ZGL.pdb), (c) thiol-inhibitor bound LytB (4H4E.pdb). |
The interaction of LytB with inhibitors has also been investigated. Figure 109 shows the partial density of vibrational states (pDOS) of LytB bound to its natural substrate 1 as well as to the inhibitors (E)-4-amino-3-methylbut-2-en-1-yl diphosphate, 2, and (E)-4-mercapto-3-methylbut-2-en-1-yl diphosphate, 3. There is reasonable agreement between calculated and experimental data and the deviations have been attributed to the presence of slightly different protein conformations in protein crystals and in solution. As already indicated by Mossbauer spectroscopy and more recently by X-ray structure studies, the inhibitor 2 coordinates with its amino group and the inhibitor 3 coordinates with its thiol group to the special site of the 4Fe-4S cluster of LytB [1,2]. However, for the interaction of the amino inhibitor 2 there are two different X-ray structures found in the pdb database. The present work shows that the vibrational properties of the enzyme complexed with inhibitor 2 are better reproduced by one of the two structures (3ZGL.pdb) indicating that this structure is present in solution.
Principal publication and authors
Isoprenoid biosynthesis in pathogenic bacteria: nuclear resonance vibrational spectroscopy provides more insight into the unusual [4Fe-4S] cluster of the LytB/IspHprotein, I. Faus (a), A. Reinhard (a), S. Rackwitz (a), J.A. Wolny (a), K. Schlage (b), H.-C. Wille (b), A. Chumakov (c), S. Krasutsky (d), P. Chaignon (e), C.D. Poulter(d), M. Seemann (e) and V. Schünemann (a), Angew. Chem. Int. Ed., 54, 12584-12587 (2015); doi: 10.1002/anie.201502494.
(a) University of Kaiserslautern (Germany)
(b) Petra III, DESY, Hamburg (Germany)
(c) ESRF
(d) University of Utah, Salt Lake City (USA)
(e) Université de Strasbourg/CNRS, UMR 7177 (France)
References
[1] A. Ahrens-Botzong et al., Angew. Chem., Int. Ed. 50, 11976−11979 (2011).
[2] I. Span et al., Angew. Chem. Int. Ed. 52, 2118 –2121 (2013).



