- Home
- News
- Spotlight on Science
- How the "green light"...
How the "green light" is given for protein translation
31-01-2011
The production of proteins is performed in all organisms by ribosomes, nanomachines that translate the genetic code inscribed in a sequence of RNA (messenger or mRNA) and then synthesise a protein from that sequence. Many mechanisms exist to make sure that the genetic code is correctly translated into a functional protein. A new crystal structure of the ribosome reveals how ribosomes check that the genetic code has been correctly recognised and trip a molecular switch to give the green light for translation to occur.
Share
DNA encodes the instructions needed to produce the proteins that perform nearly all the functions in the body, from breaking down food to muscle contraction. However, the information in DNA needs to be decoded and the first step is to transcribe the DNA sequence into a template, called messenger ribonucleic acid, mRNA. Nanomachines called ribosomes read this template and use the information contained in it to join together a precise sequence of amino acids to produce functional proteins. Ribosomes are themselves mostly made of RNA, with some protein, and are comprised of a large and a small subunit. The small subunit closes around a sequence of mRNA and it is at this point that the process of translation and peptide synthesis starts. The ribosome uses transfer RNA (tRNA) with individual amino acids attached as substrates. Each tRNA matches the three letter genetic code (codon) to a specific amino acid thus allowing, in principle, a sequence of amino acids to be attached to each other in the order dictated by the genetic code. Aminoacyl-tRNA is delivered to the ribosome by a protein called Elongation Factor-Tu (EF-Tu). This protein binds tightly to tRNA and will only release it when the genetic code has been accurately translated. Recent structures [1] have provided details on how EF-Tu interacts with the ribosome but the molecular details of the mechanism by which the green light is given for protein synthesis have remained elusive.
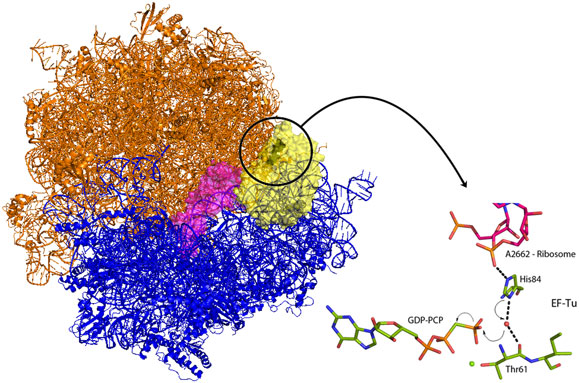
Venki Ramakrishnan’s group from the Laboratory of Molecular Biology in Cambridge (UK) has now crystallised the entire ribosome in complex with EF-Tu and aminoacyl-tRNA locked in the activated conformation and before the molecular switch has been tripped. EF-Tu will only release tRNA when a bound molecule of guanosine triphosphate (GTP) has been hydrolysed. GTP acts as a molecular switch by inducing different conformations in EF-Tu between the GTP bound form and the GDP form present after hydrolysis of GTP. The new structure demonstrates how codon recognition leads to hydrolysis of GTP and therefore allows protein synthesis to occur. An intricate interaction between tRNA, EF-Tu and the ribosome itself acts to position a charged histidine residue that activates a water molecule to hydrolyse GTP (Figure 1). Upon binding of EF-Tu and tRNA to the ribosome, and when the genetic code is correctly recognised, conformational changes are induced in the tRNA and its bound EF-Tu. These changes align EF-Tu with a region of the ribosome called the sarcin-ricin loop (SRL – so called because the poison ricin acts at this position to prevent all protein production). In the new structure, a non-hydrolysable analogue of GTP, GDP-PCP, was used to trap the complex at the instant before the molecular switch is tripped. The structure shows that the phosphate backbone of A2662 in the SRL stabilises an EF-Tu histidine residue in a position where it is perfectly positioned to activate a water molecule for nucleophilic attack on the GTP (Figure 1). Once GTP is hydrolysed, EF-Tu is able to release its bound tRNA and protein synthesis can begin and the next tRNA-EF-Tu can be bound. Elongation factors share very high sequence homology and as the SRL is an essential component of ribosomes in all kingdoms of life this newly revealed mechanism is universal to all forms of life.
As ribosomes are very large complexes (~30 nm in the largest dimension) this leads to heterogeneity in diffraction quality between crystals. The diffraction properties of hundreds of crystals needed to be examined in a synchrotron X-ray beam in order to select those that diffract to a sufficient resolution to allow the description of molecular details, such as GTP hydrolysis. This type of sample evaluation is only possible at the ESRF Structural Biology Beamlines due to their high level of automation of beam alignment and sample handling. The final data sets for these experiments were collected on beamline ID14-4, where the low divergence of the beam is extremely useful when collecting data from crystals with large unit cell dimensions.
References
[1] T.M. Schmeing et al., Science 326, 688-694 (2009).
Principal publication and authors
R.M. Voorhees, T.M. Schmeing, A.C. Kelly, and V. Ramakrishnan, The mechanism of GTP hydrolysis on the Ribosome, Science 330, 835-838 (2010).
MRC Laboratory of Molecular Biology, Cambridge (UK)
Article written by M.W. Bowler, ESRF.
Top image: Giving the green light for protein synthesis (Image credit: R. Voorhees and M. Schmeing).