- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2014
- Structural biology
- The “strong ARM” of SRP68 shapes the signal recognition particle
The “strong ARM” of SRP68 shapes the signal recognition particle
Targeted protein transport is a vital process for all cells. A priori targeting can occur while the protein is being synthesised (co-translational) or thereafter (post-translational) and each pathway depends on a dedicated machinery. In eukaryotic cells, membrane proteins and secreted proteins are co-translationally targeted to the endoplasmic reticulum (ER) by the universally conserved signal recognition particle (SRP) [1]. SRP recognises an N-terminal hydrophobic signal sequence within the nascent polypeptide chain, stalls the translation process, and upon interaction with the SRP receptor (SR), the ribosome–nascent chain complex (RNC) is docked onto the translocation channel (the Sec translocon) within the ER membrane. Here, protein biosynthesis resumes and the nascent chain is directly inserted into the ER membrane or translocates into the lumen of the ER. Eukaryotic SRP is a complex ribonucleoprotein particle consisting of six proteins assembled on a SRP RNA of 300 nucleotides (Figure 98a). SRP can be divided into two domains, the S domain, where the signal sequence binds, and the Alu domain, responsible for elongation arrest. In recent years, the molecular details of most SRP components have been determined with the beamlines at the ESRF playing a major role. However, it was only this year that structural information on SRP68, one of the largest, eukaryote specific components, became available.
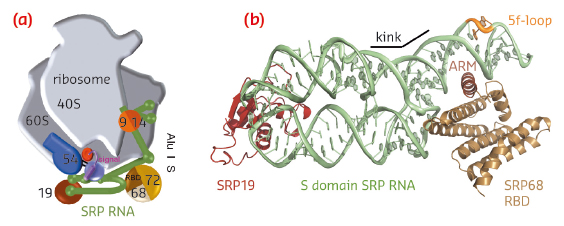 |
|
Fig. 98: Co-translational protein targeting by the signal recognition particle (SRP). a) Human SRP bound to the RNC. b) Ternary complex of SRP S domain including SRP19, SRP68-RBD, and SRP RNA. |
SRP68 forms a heterodimer with SRP72 and the SRP68 RNA-binding domain (RBD) has been previously shown within the context of the RNC to bind to a three-way junction ([2], for review see [3]). While this binding site is quite distant from the Alu domain and the signal sequence binding protein SRP54, SRP68 was found to be essential for translation-arrest and protein translocation. We have now determined the X-ray structures of the human and a fungal SRP68-RBD domain both alone and in the context of the SRP S domain. Data were collected on the tuneable beamlines ID23-1 and ID29 and initial structure determination was performed by multi-wavelength anomalous dispersion (MAD) using selenomethionine labelled protein. The high brilliance of the beamlines was essential for obtaining superior data quality from the small and weakly diffracting crystals of the protein-RNA complex.
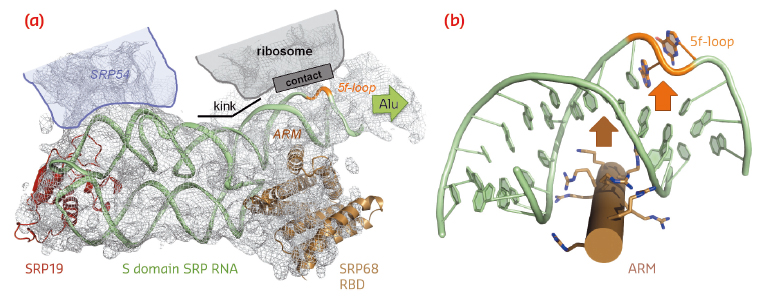 |
|
Fig. 99: SRP RNA remodelling by SRP68-RBD. a) SRP68-RBD induces a kink in SRP RNA necessary for contacting the ribosome. b) SRP68-RBD inserts an ARM into the major groove of SRP RNA and opens the 5f-loop. |
SRP68-RBD forms an unusual RNA-binding module resembling the purely α-helical tetratricopeptide repeat (TPR) fold (Figure 98b). The RBD exposes a highly positively charged surface, which tightly binds to an RNA three-way junction. While previous footprinting experiments accurately mapped the binding site on the SRP RNA, the consequences of the interaction were unexpected. SRP68-RBD remodels the SRP RNA on two levels and this remodelling elegantly explains previous structural and functional data. First, binding to the three-way junction induces a kink in S domain SRP RNA. When the new structure is fitted into the cryo-EM structure of the entire eukaryotic SRP/RNC complex, the “kinked” S domain is necessary to correctly explain the RNA density and to establish an important RNA-RNA contact between SRP and the ribosome (Figure 99a). Without SRP68 present to induce this contact, the Alu domain could not, as already described 30 years ago, function in translation-arrest. Second, one α-helix of SRP68-RBD is inserted into the major groove of SRP RNA in the 5f-loop region next to the three-way junction (Figures 98b and 99b). The major groove in RNA-helices is narrow and deep, and it needs a “strong arm” to open it. For this to happen, an arginine-rich motif (ARM) is inserted into the groove, and the 5f-loop is remodelled by bulging-out conserved unpaired nucleotides. It is these nucleotides that form the contact with ribosomal RNA (Figure 99a). A corresponding RNA-bulge in bacterial SRP RNA is essential for SRP RNA mediated GTPase activation of the SRP GTPases present in SRP and its SR [4]. GTPase activation is essential for driving the SRP cycle of co-translational targeting. Whether this mechanism also applies to the eukaryotic SRP system remains to be seen.
Principal publication and authors
J.T. Grotwinkel, K. Wild, B. Segnitz and I. Sinning, Science 344, 101-104 (2014).
Heidelberg University Biochemistry Center (BZH), Heidelberg (Germany)
References
[1] P. Grudnik, G. Bange and I. Sinning, Biol Chem 390, 775-782 (2009).
[2] M. Halic et al., Nature 427, 808-814 (2004).
[3] K. Wild, M. Halic, I. Sinning and R. Beckmann, Nat Struct Mol Biol 11, 1049-1053 (2004).
[4] G. Bange and I. Sinning, Nat Struct Mol Biol 20, 776-780 (2013).



