- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2014
- Structural biology
- Structural elucidation of MADS domain transcription factor SEPALLATA3
Structural elucidation of MADS domain transcription factor SEPALLATA3
Virtually all terrestrial habitats are dominated by angiosperms, or flowering plants. However, angiosperms are a relatively recent step in evolution. Their success in colonising new habitats and superseding other species is due to the advent of a novel reproductive structure – the flower. Flowers are composed of four different organs: the sepals which protect the developing bud, the petals which are the showy organs that act as pollinator attractors, the stamen which are the male organs containing the pollen and the carpels which are the female organs and contain the ovules. The flower unites the male and female organs into one structure and encloses the seed. Flowering plants are not only the dominant type of land plants, but are also the primary source of food and habitat for all animals, including humans. The evolution of flowers has been the subject of speculation from the time of Charles Darwin who termed the dominant rise and diversification of flowering plants as “an abominable mystery” due to the lack of a smooth transition from non-flowering to flowering plants in the fossil record. With the sequencing of multiple genomes from basal angiosperms, gymnosperms and higher flowering plants, certain gene families that play a central role in the development and evolution of the flower have been identified. Our research focuses on one such family of high-level regulators called the MADS transcription factor (TF) family, that orchestrates flower development. We are interested in understanding the molecular mechanisms of the MADS family and how these proteins are able to control complex reproductive functions.
Unlike animals which have only a few MADS family members, plants have greatly expanded and diversified the family during evolution. MADS TFs play significant roles in all aspects of plant life from seed germination to fruit and flower development. One of the most central members of the MADS family is SEPALLATA3 (SEP3). SEP3 is able to form over 50 different complexes with other MADS proteins and has roles in virtually all aspects of plant reproduction including the establishment of the floral meristem, the development of all floral organs and fruit ripening [1]. SEP3-containing protein complexes are postulated to bind DNA at multiple binding sites resulting in DNA loops that are important for downstream gene expression and/or repression [2]. Some of our recent work investigated the molecular mechanisms of SEP3 function. Using a combination of structural techniques, including X-ray crystallography and atomic force microscopy (AFM), we determined how SEP3 is able to form different protein-protein complexes and directly demonstrated DNA looping following SEP3 tetramer formation.
 |
|
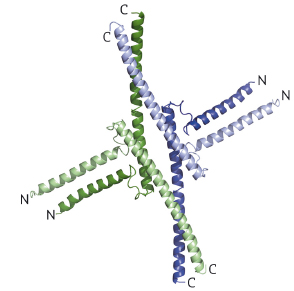
Fig. 110: SEP3 tetramer depicted as a cartoon with each monomer coloured differently. For each monomer, the N and C termini are labelled. |
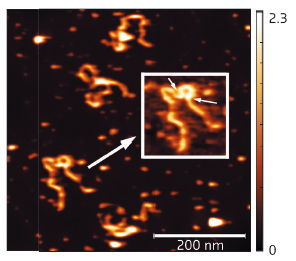
The 2.5 Å resolution crystal structure of the SEP3 oligomerisation domain was determined from diffraction data obtained at beamline ID14-4 (now ID30B). This is the first structure for a plant MADS transcription factor and demonstrates novel dimerisation and tetramerisation interfaces formed by amphipathic α-helices. Figure 110 shows that the protein monomer folds into two amphipathic α-helices separated by a rigid kink region, that the dimer is formed via hydrophobic interactions from helix 1 and helix 2 and that the tetramerisation interface is more limited, relying on hydrophobic residues contributed by the C-terminal portion of helix 2. Mutagenesis studies have confirmed the importance of these residues to the tetramerisation of the protein. To study the full-length protein, AFM was performed in collaboration with F. Comin and L. Costa of the ESRF Surface Science Laboratory. These experiments directly demonstrate SEP3-DNA binding. Figure 111 shows a SEP3 tetramer binding two adjacent DNA-binding sites leading to the formation of a DNA loop. This ability is lost when the C-terminal portion of helix 2 is deleted.
 |
|
Fig. 111: AFM results showing full length SEP3 in complex with a 1-kb DNA fragment containing two SEP3 binding sites. The inset clearly shows that SEP3 tetramer binding to two adjacent DNA-binding sites (indicated by arrows) leads to formation of a DNA loop. |
These results provide the first structural basis for MADS domain transcription factor function. The novel dimerisation and tetramerisation interfaces present two independent protein-protein interaction surfaces, allowing the proper binding of many different partners for both dimer and tetramer formation. We are currently developing and testing predictive models for MADS domain TF function based on our structural studies of SEP3 using an integrated biophysical and genetic approach. As a long-term goal, these studies will help us to tune the function of MADS TFs in plants and to alter different reproductive pathways and structures in a predictable manner.
Principal publication and authors
S. Puranik (a), S. Acajjaoui (a), S. Conn (b), L. Costa (a), V. Conn (b), A. Vial (a), R. Marcellin (a,c), R. Melzer (d), E. Brown (a), D. Hart (e), G. Theißen (d), C.S. Silva (f), F. Parcy (f), R. Dumas (f), M. Nanao (g,h) and C. Zubieta (a,f,i), The Plant Cell 26, 3606-3615 (2014).
(a) ESRF
(b) SA Pathology and the University of South Australia, Adelaide (Australia)
(c) Faculté des Sciences de Montpellier (France)
(d) Friedrich Schiller University, Jena (Germany)
(e) Université Grenoble Alpes, CNRS, Integrated Structural Biology Grenoble, UVHCI, Unité Mixte Internationale 3265, UMS 3518 Grenoble (France)
(f) LPCV, Grenoble (France)
(g) EMBL, Grenoble (France)
(h) UVHCI, Université Grenoble Alpes-EMBL-CNRS, Grenoble (France)
(i) CEA, DSV, iRTSV, Grenoble (France)
References
[1] R.G.H Immink, I.A. Tonaco, S Folter, A Shchennikova, A.DJ. Dijk, J. Busscher-Lange and J.W. Borst, Genome Biology 10, R24 (2009).
[2] R. Melzer, W. Verelst and G. Theissen, Nucleic Acids Res. 37, 144-157 (2009).



