- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2002
- Macromolecular Crystallography
- Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates Along the Reaction Pathway
Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates Along the Reaction Pathway
Tuberculosis is the primary cause of human mortality from infectious agents, with one-third of the world's population infected. In the absence of an effective vaccine, the need for new drugs against tuberculosis has become a priority. Kinases responsible for the activation of nucleoside to nucleotide triphosphates, the building blocks of RNA and DNA, represent promising targets. Of special interest is thymidylate kinase (TMPK), an enzyme essential to cell survival, catalysing the phosphorylation of deoxythymidine-5'-monophosphate (TMP) to deoxythymidine-5'-diphosphate (TDP). Like other TMPKs, Mycobacterium tuberculosis TMPK (TMPKMtub) is a homodimer in solution (Mm = 44 kDa) with each monomer being composed of nine ![]() -helices surrounding a five-stranded ß-sheet core [1]. The active site includes the usual P-loop (a phosphate binding loop) and LID region (a highly flexible stretch of residues covering the ATP binding site). However, subtle sequence deviations from other TMPKs [1] confer unique properties to TMPKMtub, suggesting that inhibitors could specifically target this enzyme.
-helices surrounding a five-stranded ß-sheet core [1]. The active site includes the usual P-loop (a phosphate binding loop) and LID region (a highly flexible stretch of residues covering the ATP binding site). However, subtle sequence deviations from other TMPKs [1] confer unique properties to TMPKMtub, suggesting that inhibitors could specifically target this enzyme.
In order to clarify the phosphorylation mechanism employed by TMPKMtub, we have used kinetic crystallography and investigated the sequence of structural changes that take place upon the binding of substrates and catalysis.
Crystallisation of TMPKMtub using sodium malonate as precipitating agent and initiation of catalysis in the crystal by diffusion of ATP allowed us to elucidate the structures of (i) the apo-TMPKMtub (Figure 8a), (ii) the complex TMPKMtub-TMP (Figure 8b), (iii) the complex TMPKMtub-TMP-Mg2+ (Figure 8c) and (iv) the complex TMPKMtub-TDP-ADP-Mg2+ (Figure 8e). In addition, crystals grown in the presence of ammonium sulphate show a magnesium ion and a sulphate ion in the ATP binding site and provide a good mimic of the complex TMPKMtub-TMP-ATP-Mg2+ (Figure 8d, [1]).
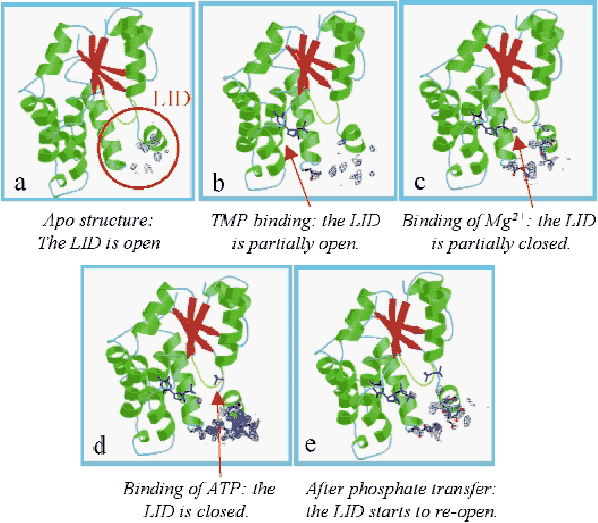 |
|
Fig. 8: Omit maps calculated by omitting 17 residues (149-165) in the LID region and contoured at 1.2 |
Overall, a series of snapshots along the reaction pathway is obtained, revealing a closure of the active site in the process of going from an empty to a fully-occupied state. This suggests an induced-fit mechanism typical of NMPKs. These results also demonstrate that in TMPKMtub the LID closure is specifically coupled to the transient binding of an unusual magnesium ion coordinating TMP in the active site. To clarify the role of this ion, crystals were soaked in ATP at acidic pH, where the enzyme is inactive (Figure 9a). In such conditions there is no Mg2+ bound to the enzyme. The overall geometry of the TMP binding site is disrupted and no formation of TDP occurs (Figure 9b), revealing that the presence of the metal ion is indispensable in bringing together the ![]() -phosphate of TMP and the
-phosphate of TMP and the ![]() -phosphate of ATP for efficient phosphoryl transfer.
-phosphate of ATP for efficient phosphoryl transfer.
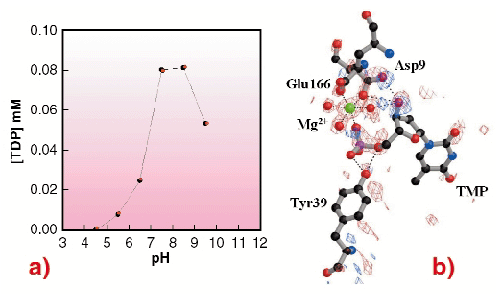 |
|
Fig. 9: (a) pH dependence curve of the TMPKMtub catalytic activity. The TDP production (on the vertical axis) is represented as a function of pH (horizontal axis); (b) TMPKMtub active site. Difference electron density map contoured at ± 3.5 |
The essential role of Mg2+ in driving catalysis in TMPKMtub suggests that structure-based design of inhibitors for the enzyme should focus on possibilities to destabilise this ion. The low pH structure shows that the 3'-hydroxyl group of the TMP substrate participates in binding Mg2+. Potential inhibitors designed to contain a 3'-hydroxyl group substituent without hydrogen bonding capability may therefore prevent the binding of the metal ion and abolish the catalytic activity of TMPKMtub.
Reference
[1] I. Li de la Sierra, H. Munier-Lehmann, A.M. Gilles, O. Bârzu and M. Delarue, J. Mol. Biol. 311, 87-100 (2001).
Principal Publication and Authors
E. Fioravanti (a,b), A. Haouz (c), T. Ursby (d), H. Munier-Lehmann (c), M. Delarue (c) and D. Bourgeois (a,b), to be published.
(a) LCCP, UMR 9015, IBS, Grenoble (France)
(b) ESRF
(c) Institut Pasteur, Paris (France)
(d) MAX-lab, Lund University, Lund (Sweden)



