- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2005
- X-ray Absorption and Magnetic Scattering
- X-ray Absorption Studies on the N-terminal Copper-binding Region of Haemophilus ducreyi Cu,Zn Superoxide Dismutase
X-ray Absorption Studies on the N-terminal Copper-binding Region of Haemophilus ducreyi Cu,Zn Superoxide Dismutase
Copper is an essential element for all aerobic organisms which incorporate this metal ion in several important enzymes. One of these enzymes is Cu,Zn superoxide dismutase (Cu,ZnSOD), which protects cells from the toxic effects of reactive oxygen intermediates by converting the superoxide radical into hydrogen peroxide and molecular oxygen. In bacteria Cu,ZnSOD is located in extracytoplasmic compartments where it plays an important role in protecting microorganisms from exogenous sources of superoxide [1]. The Cu,ZnSOD from a subset of Gram-negative pathogens possess divalent metal binding N-terminal extensions, which presumably favours the uptake of the enzyme’s prosthetic metals in environments where their concentration is very low [2]. In particular, the N-terminal extension of the Cu,ZnSOD from Haemophilus ducreyi, the causative agent of a genital ulcerative disease known as chancroid, shows particularly interesting features (HGDHMHNHDTKMDTMSKDMMSM). The N-terminus of this motif, which contains a cluster of four histidines interspersed with other residues, is very similar to the transition metal binding regions already identified in other proteins that are able to bind Ni(II), Zn(II), or Cu(II) [2]. The second half of the domain contains an unusual methionine-rich sequence that resembles the Cu(I)-binding domains observed in several proteins involved in copper homeostasis in prokaryotic and eukaryotic cells [3,4]. These observations suggest that the N-terminal domain of H. ducreyi Cu,ZnSOD could allow the efficient binding of Cu(II) and Cu(I) at the histidine-rich and methionine-rich sequences, respectively.
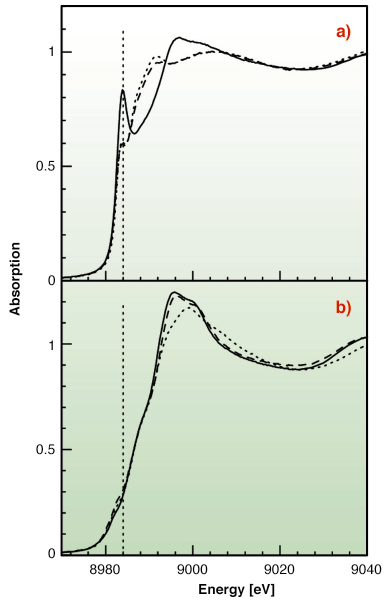 |
|
Fig. 129: (a) Cu K-edge XANES spectra for Cu(I) with Met-rich (dotted line), His-rich (solid line), and an equimolar mixture of the two peptides (dashed line) in water solution. (b) Cu K-edge XANES spectra for Cu(II) with Met-rich (dotted line), His-rich (solid line), and an equimolar mixture of the two peptides (dashed line) in water solution. |
To test this hypothesis X-ray absorption spectroscopy experiments have been carried out on peptides corresponding to the two metal binding regions. Cu K-edge XAS spectra were collected in fluorescence mode at the BM30-B (FAME) beamline on samples with Cu concentrations of 2.5 and 1.25 mM. Our results indicate that both sequences can bind either Cu(II) or Cu(I) as shown in Figure 129. However, competition experiments demonstrate that Cu(II) is preferred by histidine residues belonging to the first half of the motif, while the methionine-rich region preferentially binds Cu(I) via the interaction with three methionine sulfur atoms as shown in Figure 130. These findings demonstrate that the N-terminal domain of H. ducreyi Cu,ZnSOD is an unusual metal trapping peptide able to bind Cu(I) or Cu(II) at different sites, possibly favouring an efficient copper uptake under copper starvation. Moreover, alterations in the copper coordination environment mediated by changes in the copper redox state could play a role in favoring metal transfer from the N-terminal domain to the active site.
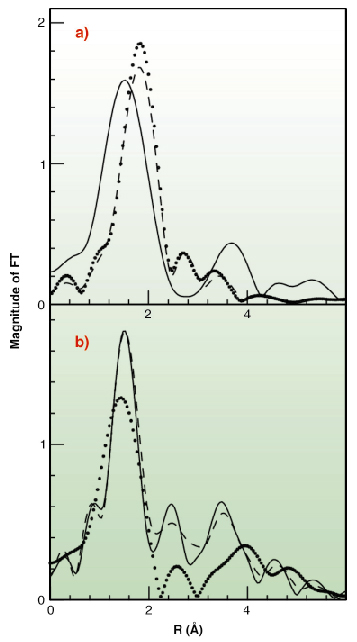 |
|
Fig. 130: Fourier Transforms of the k2-weighted EXAFS experimental data. (A) Cu(I) complexes (B) Cu(II) complexes. Data are shown as dotted lines (Met-rich peptide), solid line (His-rich peptide), and dashed line (equimolar mixture of the two peptides). |
In conclusion, we suggest that the juxtaposition of two sequence motifs potentially involved in the binding of Cu(I) and Cu(II) in the N-terminal domain of H. ducreyi Cu,Zn-SOD could have a double role: to facilitate copper acquisition in environments poor of this metal ion and to assist Cu(I) transfer from the high-affinity Met-rich motif to the enzyme active site.
References
[1] A. Battistoni, A. Biochem. Soc. Trans. 31, 1326-1329 (2003).
[2] A. Battistoni, F. Pacello, A. P. Mazzetti, C. Capo, S. J. Kroll, P. Langford, A. Sansone, G. Donnarumma, P. Valenti, G. Rotilio J. Biol. Chem. 276, 30315-30325 (2001).
[3] D.L. Huffman, J. Huyett, F.W. Outten, P.E. Doan, L.A. Finney, B.M. Hoffman, T.V. O’Halloran, Biochemistry, 41, 10046-10055 (2002).
[4] K. Peariso, D.L. Huffman, J.E. Penner-Hahn, T.V. O’Halloran J. Am. Chem. Soc., 125, 342-343 (2003).
Principal publication and Authors
P. D’Angelo (a), F. Pacello (b), G. Mancini (a), O. Proux (c), J.-L. Hazemann (d), A. Desideri (b,e), A. Battistoni (b,e) Biochemistry 44, 13144-13150 (2005).
(a) Dipartimento di Chimica Università di Roma “La Sapienza” (Italy)
(b) Dipartimento di Biologia Università di Roma Tor Vergata (Italy)
(c) Laboratoire de Géophysique Interne et Tectonophysique, UMR CNRS, Université Joseph Fourier, Saint-Martin- d’Hères (France)
(d) Laboratoire de Cristallographie, CNRS, Grenoble (France)
(e) INFM, Università di Roma Tor Vergata (Italy)



