- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2007
- Structural Biology
- Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G
Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G
Entry of enveloped viruses into host cells requires binding of the virus to one or more receptors present at the host cell surface followed by fusion of the viral envelope with a cellular membrane. Activation of the fusion capacity involves large structural rearrangements of fusogenic glycoproteins upon interaction with specific triggers (e.g. low pH or cellular receptors). These conformational changes result in the exposure of a fusion peptide or fusion loops, which then interact with one or both of the participating membranes resulting in their destabilisation and merging [1].
Rhabdoviruses are enveloped viruses with a single-stranded non-segmented RNA genome of negative polarity. Vesicular stomatitis virus (VSV) is the prototype of the family. Its membrane contains a single transmembrane glycoprotein (G) that is involved in both receptor recognition and membrane fusion. Fusion is triggered by low pH-induced structural rearrangements during which G shifts from a prefusion native state to a postfusion conformation. In sharp contrast to fusogenic glycoproteins from other viral families, this low-pH induced conformational change is reversible: there is a pH dependent equilibrium between the different conformations of G that is shifted toward the postfusion state at low pH [2].
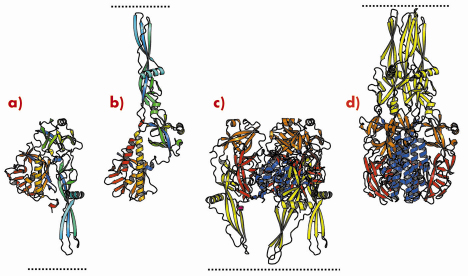
We have previously determined the low pH, post-fusion, 3D structure of the VSV G ectodomain generated by limited proteolysis of the virions. It remained unclear to what extent the pre- and post-fusion conformations differ and thus we have also determined the structure of the prefusion form of VSV G. Comparison of the pre- and postfusion structures of VSV G reveals a dramatic reorganisation of the molecule that probably constitutes the largest reversible conformational change ever described for a protein (Figure 67). During the conformational change, three domains of G (out of four) retain their tertiary structure. Nevertheless, they undergo large rearrangements in their relative orientation due to secondary structure changes in hinge regions. The fourth domain, that is involved in the trimerisation of both the pre- and the post-fusion conformation, undergoes a major refolding event that includes large changes in its secondary structure (Figure 67c and 67d).
 |
|
Fig. 67: Ribbon representation of the G protomer (a, b) and trimer (c, d) in both prefusion (a, c) and postfusion (b, d) conformations. Protomers are coloured by residue number in a gradient from blue (N terminus) to red (C terminus). Trimers are coloured by domain (lateral domain: red, trimerisation domain: blue, pleckstrin homology domain: orange, fusion domain: yellow) with the C-terminus in magenta. Pre- and post-fusion forms are superimposed on the rigid blocks made of the lateral domain and the invariant part of the trimerisation domain. The lipid bilayer, in which the transmembrane domains are anchored, is indicated by the dotted line. |
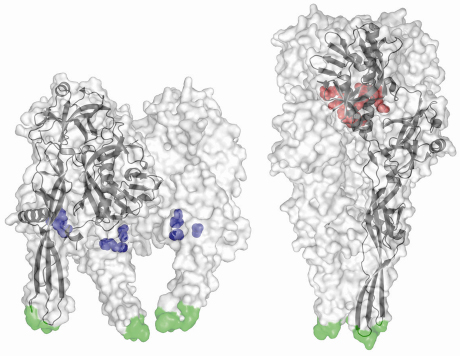
Furthermore, we have previously identified acidic amino acid residues that are brought together in the central six helix bundle in the post-fusion (low pH) state [3]. In this conformation, they are protonated and participate in hydrogen bonds. The deprotonation of these residues at higher pH will induce strong repulsive forces that destabilise the trimer and initiate the conformational change back to the prefusion state (in which these residues are solvent exposed). Conversely, the prefusion structure revealed a conserved cluster of three histidines that could play the role of a pH sensitive molecular switch. Indeed, low pH–induced protonation of these residues leads to a cluster of positive charges that might trigger the movement of the fusion domain toward the target membrane (Figure 68).
 |
|
Fig. 68: Structures of the VSV-G ectodomain in prefusion (left) and postfusion (right) conformations. For each structure, the trimer is displayed as a semitransparent white surface with the fusion loops in green and a protomer is also drawn in black in ribbon represention. Despite its magnitude, the pH-triggered conformational change is fully reversible, with protonation of the histidine side chains (shown in blue) starting the forward transition and deprotonation of the aspartates and glutamates (red) driving the backward transition. |
Finally, it is noteworthy that in the case of rhabdoviruses, the pH dependent equilibrium between pre and post-fusion conformations of G indicates that the energy released during the structural transition of one trimer cannot be sufficient to overcome the energetic barrier and drive the fusion reaction. Indeed, the minimal number of spikes involved in the formation of a RV fusion complex has been estimated to be about 15 [2]. Interestingly, the crystalline form (P622) of the prefusion form reveals the ability of G to adopt a planar p6 lattice. We propose that such a local hexagonal lattice at the viral surface might organise the glycoproteins in an optimal manner for a concerted conformational change that can be used to overcome the high energy barrier encountered during fusion.
References
[1] W. Weissenhorn, A. Hinz, Y. Gaudin, FEBS Lett. 581, 2150 (2007).
[2] S. Roche, Y. Gaudin, Virology 297, 128 (2002).
[3] S. Roche, S. Bressanelli, F.A. Rey, Y. Gaudin, Science 313, 187 (2006).
Principal publication and authors
S. Roche, F.A. Rey, Y. Gaudin, S. Bressanelli, Science 315, 843 (2007).
UMR-CNRS 2472, UMR-INRA 1157, Laboratoire de Virologie Moléculaire et structurale, Gif sur Yvette (France)



