- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2008
- Soft condensed matter
- Directed motion of proteins along tethered polyelectrolytes
Directed motion of proteins along tethered polyelectrolytes
The adsorption and immobilisation of proteins from aqueous solutions onto solid surfaces is a topic of considerable interest in both soft matter and biochemical research [1]. Many biologically important processes are interfacial in nature and very often, charged and uncharged polymers attached to surfaces are used to tune the interaction with the proteins. Various biotechnological processes require immobilisation of enzymes with full retention of their biological activity. Reversible adsorption of proteins onto charged surfaces is used in protein purification by ion exchange chromatography and in many natural processes such as cell adhesion. Nevertheless, unspecific adsorption of proteins must be suppressed in many practical applications in order to prevent foreign body reactions to materials from a host. Little is known about the kinetics of protein adsorption and the self-organisation of biomolecules with tethered polymer chains at a molecular level.
 |
|
Fig. 45: Sketch of the uptake of proteins (red spheres) by spherical brushes consisting of a solid core (grey sphere) and grafted polyelectrolyte chains (wavy lines). |
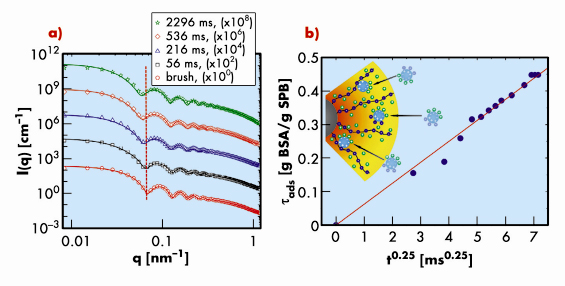
Here we report on the first study of the motion of proteins (bovine serum albumin, BSA) in a layer of tethered polyacrylic acid chains on colloidal polystyrene particles (Figure 45) using time-resolved small-angle X-ray scattering (TR-SAXS) combined with stopped-flow rapid mixing at beamline ID02. The pH value of the aqueous solution was kept well above the isoelectric point of the charged proteins and the ionic strength in the solution was adjusted to 7 mM to ensure full uptake of proteins by the spherical polyelectolyte brushes (SPB). The driving force is the counterion release mechanism, i.e., the exchange of confined counterions with charged proteins. Figure 46a displays the typical evolution of the scattering intensity, I(q), as a function of time, t. The gradual uptake of protein can be followed directly from the shift of the side maxima to smaller scattering vectors q with increasing time (see the dotted vertical line). The continuous lines indicate the calculated intensities using the radial electron density profile of SPB loaded with BSA and the screened Coulomb interaction between the SPB obtained from a reference interaction site integral equation theory. The TR-SAXS data revealed that the uptake of the proteins into the brush layer is relatively fast and the final state is reached within 3 seconds. However, the time needed by a single protein molecule to diffuse freely through the tethered layer is less than a millisecond. This means that the motion of the proteins in the brush layer has been slowed significantly. From the radial electron density profiles, the total amount of adsorbed protein per SPB (![]() ads) at a given time can be derived. Figure 46b depicts that tads increases with time as t1/4. For unhindered diffusive motion of proteins, the mean-square-displacement is expected to increase with time as t1/2. The observed sub-diffusive behaviour is due to a balance of frictional and external forces acting on the proteins as shown schematically in the inset of Figure 46b.
ads) at a given time can be derived. Figure 46b depicts that tads increases with time as t1/4. For unhindered diffusive motion of proteins, the mean-square-displacement is expected to increase with time as t1/2. The observed sub-diffusive behaviour is due to a balance of frictional and external forces acting on the proteins as shown schematically in the inset of Figure 46b.
 |
|
Fig. 46: a) TR-SAXS intensities, I(q), as a function of time, t. For clarity, successive upper curves have been shifted by a factor 100 (as indicated in the legend). The continuous lines represent the model. b) The time dependence of the amount of adsorbed protein obtained from the radial electron density profiles derived from the fits in (a). The inset illustrates the directed motion of proteins through the brush layer. |
In summary, we have demonstrated that protein adsorption can be monitored directly with high temporal and spatial resolution on surface-modified colloidal spheres using TR-SAXS. The uptake of charged proteins by spherical polyelectrolyte brushes follows a sub-diffusive behaviour. A similar directed motion of proteins and other biomolecules through biological membranes could play a pivotal role in their self-assembly.
Principal publication and authors
K. Henzler (a), S. Rosenfeldt (a), A. Wittemann (a), L. Harnau (b), S. Finet (c), T. Narayanan
(c), and M. Ballauff (a), Phys. Rev. Lett. 100, 158301 (2008).
(a) Physikalische Chemie I, University of Bayreuth (Germany)
(b) MPI Stuttgart and ITAP, University of Stuttgart (Germany)
(c) ESRF
References
[1] Proteins at Solid-Liquid Interfaces, P. Dejardin (Ed), Springer (2006).



