- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2010
- Structural biology
- A prokaryotic transcription regulation mechanism involving allosteric coupling between intrinsically disordered domains
A prokaryotic transcription regulation mechanism involving allosteric coupling between intrinsically disordered domains
Regulation of transcription is a fundamental process common to all kingdoms of life. Transcription regulation in eukaryotes, however, is generally considered to be significantly more complex than the corresponding mechanisms used by prokaryotes. In particular, eukaryotic transcription regulation makes an abundant use of intrinsic disorder, leading to the concept that eukaryotic transcription regulators are malleable machines that adapt themselves to recognise different targets in response to environmental or temporal cues. The presence of intrinsic disorder in prokaryotes is less well pronounced and was not generally thought to be crucial for transcription regulation. Transcription regulation in prokaryotes is known mostly via a limited number of well-studied examples, such as the choice between a lytic or a lysogenic cycle for phage λ.
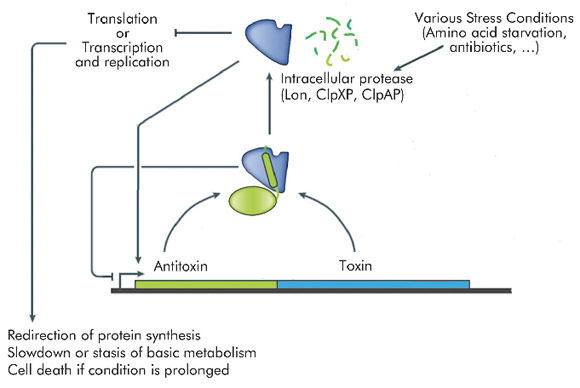
Many transcriptional mechanisms in prokaryotes remain unexplored. An interesting example is regulation by conditional co-operativity in toxin-antitoxin (TA) modules. TA modules are stress response elements where a growth regulator or inducer of altruistic cell death (the toxin) is kept under control of a second protein (the antitoxin) which couples toxin neutralisation to transcription regulation (Figure 106). TA modules are auto-regulated. The antitoxin contains a DNA binding domain flanked by an intrinsically disordered toxin-binding domain. DNA binding by the antitoxin is modulated by the toxin. In the absence of the toxin, its affinity for the operator DNA is insufficient for efficient repression. Low ratios of toxin increase this affinity significantly, leading to tight repression. At higher ratios of toxin to antitoxin, the affinity of the repressor complex weakens again and transcription is fired up. This unique mechanism, termed "conditional co-operativity", allows the cell to maintain a delicate balance between toxin and antitoxin without risking accidental toxin activation, which may lead to cell death.
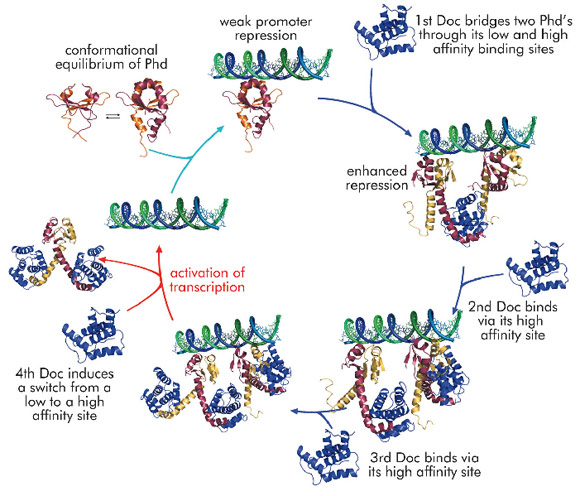
Using the phd/doc toxin-antitoxin module on bacteriophage P1 as a model system, we were able to propose a consistent molecular model that explains conditional co-operativity (Figure 107). We demonstrated experimentally that the transcription factor Phd contains a significant amount of disorder in both its domains and that this disorder is crucial for the regulation of the operon. Phd exists in an equilibrium between two conformational states: a DNA binding competent state that is highly ordered and an inactive state that contains significant disorder. Binding of Doc to the intrinsically disordered
C-terminal domain of Phd structures the N-terminal DNA binding domain of Phd. Thus, allosteric coupling occurs between two disordered biding sites, a phenomenon predicted on theoretical grounds but never before observed experimentally.
The crystal structure of the repressing Phd/Doc complex shows a Doc monomer sandwiched between two Phd dimers. In the non-repressing complex, a different architecture is observed with a Phd dimer sandwiched between two Doc monomers. The potential to create stoichiometrically different complexes coupled to a switch between a low and a high affinity interaction allowed us to propose a model that explains regulation by conditional co-operativity, where Doc acts as a co-repressor at low Doc to Phd ratios and as a de-repressor at high Doc to Phd ratios.
Principal publication and authors
A. Garcia-Pino (a,b), S. Balasubramanian (c), L. Wyns (a,b), E. Gazit (d), H. De Greve (a,b), R.D. Magnuson (c), D. Charlier (e), N.A.J. Van Nuland (a,b) and R. Loris (a,b), Cell 142, 101-110 (2010).
(a) Dept. of Molecular and cellular Interactions, Flanders Institute for Biotechnology (VIB), Brussels (Belgium)
(b) Structural Biology Brussels, Vrije Universiteit Brussel (Belgium)
(c) Department of Biological Sciences, University of Alabama in Huntsville (USA)
(d) Department of Molecular Microbiology and Biotechnology, Tel Aviv University (Israel)
(e) Erfelijkheidsleer en Microbiologie, Vrije Universiteit Brussel (Belgium)