- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2014
- Structural biology
- Tracking the bacterial immune system
Tracking the bacterial immune system
Alpha-2-macroglobulins (A2Ms) are plasma proteins that trap and inhibit a broad range of proteases and are major components of the eukaryotic innate immune system. Their main functions include clearing pathogenic or parasitic proteases from circulation, as well as blocking proteases that participate in inflammatory and blood clotting events. A2Ms are composed of ~180 kDa subunits that generally associate into oligomeric forms. The human form, for example, is a tetramer (thus, a 720 kDa protein). Despite it being believed that these molecules existed uniquely in metazoans, A2M-like proteins were recently identified in pathogenically invasive bacteria as well as in species that colonise plants, fish, and insects [1], suggesting that bacteria could also require ‘protection’ from proteases secreted by the target host or by other organisms competing for nutrients or space.
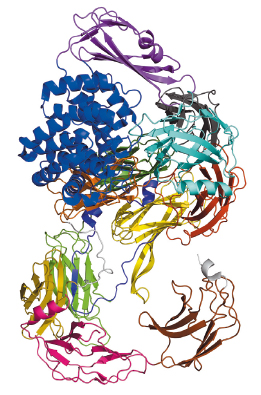 |
|
Fig. 112: Schematic arrangement of the S. typhimurium A2M, which comprises over 1600 residues that fold mostly into β-sheet-carrying domains. The only exception is the thioester domain (TED), which is mostly helical (blue), and harbours the thioester bond that attacks target proteases. |
Bacterial A2Ms are monomeric molecules located in the periplasm of the cell, thus right underneath the outer membrane, where they are believed to provide protection to the cell upon membrane breach [2]. This occurs by trapping external proteases through a covalent interaction with a thioester bond buried within a hydrophobic environment inside the inactive macroglobulin. In the test tube, activation of the thioester can be mimicked by a small molecule called methylamine. Our paper reports the crystal structures and the characterisation of the alpha-2 macroglobulin from the human pathogen Salmonella typhimurium (StA2M) in different states of thioester activation. Diffraction data were collected at beamlines ID29 and ID23-2, and solution scattering data at beamline BM29. The crystal structures revealed thirteen domains (Figure 112), whose arrangement displays high similarity to proteins involved in eukaryotic immune defense, such as complement C3. The crystal structure of (StA2M) in complex with methylamine confirms that the small molecule can attack the thioester bond without engendering conformational modification of the macroglobulin. Interestingly, the thioester motif is stabilised by a structural lock provided by Tyr1175, a residue which is present only in A2Ms of bacterial species. The necessity for this ‘lock’, which guarantees that the thioester remains entrenched (and protected from hydrolysis) within the macroglobulin in the absence of a target protease, is potentially linked to the fact that bacterial A2Ms are monomeric, and thus structurally simplistic, when compared to their oligomeric eukaryotic counterparts. Human A2M, for example, entraps target proteases in a central cage-like structure formed by its four monomers. In StA2M a Y1175G mutation opens this lock. Here, the thioester region is unprotected, the covalent thioester bond is broken, and the macroglobulin is unable to trap proteases. Our structural and mechanistic studies thus not only pinpoint how bacteria can inhibit protease attack during infection or colonisation, but also indicate that bacteria have developed a rudimentary innate immune system whose mechanism mimics that of eukaryotes.
Principal publication and authors
S.G. Wong (a), and A. Dessen (a,b), Nature Comm. 5, 4917 (2014).
(a) Institut de Biologie Structurale (IBS), CNRS, CEA, Univ. Grenoble (France)
(b) Brazilian National Laboratory for Biosciences (LNBio), Campinas (Brazil)
References
[1] A. Budd, S. Blandin, E. Levashina and T.J. Gibson, Genome Biol 5, R38 (2004).
[2] N. Doan and G.W. Gettins, J Biol Chem 283, 28747-28756 (2008).



