- Home
- News
- General News
- Visualising how...
Visualising how cell signalling takes place in microorganisms
18-05-2017
An international collaboration has unveiled the mechanisms of the crucial process of signalling through membrane receptors.
Share
Cell signalling is part of a communication process that governs the basic activities of cells and coordinates all their actions. Cells can detect what's going on around them, and respond in real-time to cues from their neighbours and environment.
It’s through membrane-associated signalling systems that bacteria and other microorganisms perceive, and react to, their environment. A major class of such systems is the sensor histidine kinases, which are two-component signalling systems (TCS). Tens of thousands of TCS are known, with many of them being essential for cell growth, survival and pathogenicity. TCS are therefore potential targets for antimicrobial drugs, such as antibiotics or antifungals. However, until today, despite extensive and world-wide research on the structural mechanisms of transmembrane signalling by TCS, they remained poorly understood at an atomic level.
 |
|
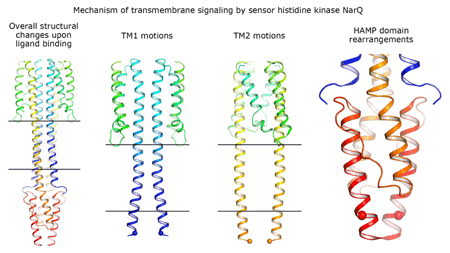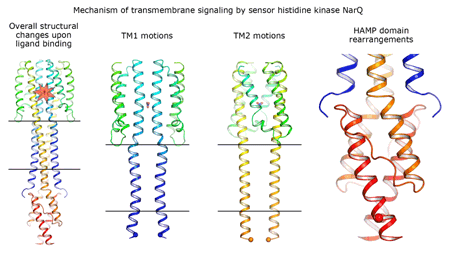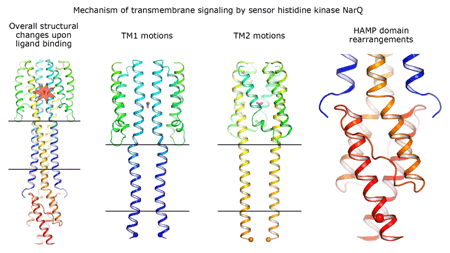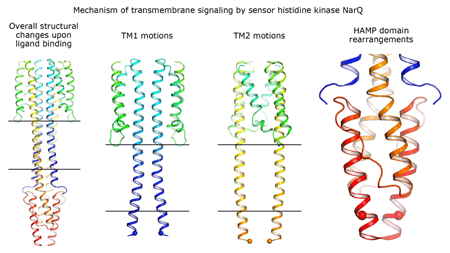
Cartoon representation of the crystal structure of the bacterial NarQ nitrate/nitrite sensor histidine kinase. Binding of the ligand causes a lever-like conformational change resulting in dissociation of the signalling helix. Copyright: Ivan Gushchin, Moscow Institute of Physics and Technology. |
Using state-of the-art membrane protein crystallisation techniques and X-ray diffraction data collected at beamline ID23-1 at the ESRF, an international team of scientists has now unveiled crystal structures of bacterial NarQ nitrate/nitrite sensor histidine kinase from the. Their results are published in Science today. The crystallised fragment of NarQ contains the periplasmic sensor domain, the TM domain and the cytoplasmic HAMP domain. A comparison of crystal structures obtained with and without nitrate bound to the sensor domain reveals the mechanism of signal transduction, showing how small, extracellular signals can be transmitted over distances of 100 Å or more. Igor Melnikov, researcher at the ESRF, explains that “all bacteria share common mechanisms in signal transduction across the membrane, so what we learned from E. coli is applicable to other bacterial species”.
Valentin Gordeliy, scientist at the IBS, Institut de Biologie Structurale (CEA, CNRS, University Grenoble Alpes) and corresponding author of the study, is hopeful that this study on NarQ will allow progress in this research: “We believe that the results reported here will help in understanding a broad range of cellular signalling processes and could contribute to the development of novel antimicrobial treatments targeting TCS”.
New method
The technique (I-SAD) used to solve the crystal structures, consisted of soaking crystals in sodium iodide, then collecting anomalous dispersion data for use in structure solution. Based on a common feature of all membrane proteins – the clustering of positively charged and large hydrophobic amino acid residues close to membrane interfaces, the team postulated that I-SAD could be used to solve the crystal structures of all classes of membrane proteins. To this end, and in a separate work, the team demonstrated the suitability of I-SAD in the solution of the crystal structures of four targets representing different classes of membrane proteins, including a human G protein–coupled receptor (GPCR). In each case, structure solution was straightforward, suggesting, according to Gordon Leonard, head of the Structural Biology Group at the ESRF, that I-SAD “is a fast, easy and potentially universal technique for the solution of membrane protein crystal structures”.
International and local flavour
The team collaborating of these projects includes groups from many institutes from all over the world: The Moscow Institute of Physics and Technology (Russia), the Institut de Biologie Structurale (France), the Research Centre Jülich (Germany), the University of Aachen (Germany), the EMBL, the University Grenoble Alpes (France), CNRS (France), CEA (France), Inria (France), Heinrich-Heine University (Germany) and the ESRF. “This work shows the strength of structural biology on the Grenoble EPN campus. Here we have specialists in many different domains and with links to many other research institutes. This helps difficult projects such as this one to succeed”, explains Leonard. “The collaboration also highlights the benefit of Russian participation in the ESRF and shows that we can go a long way working together”.
 |
|
Local members of the team, Valentin Gordeliy (IBS), Igor Melnikov (ESRF) and Alexander Popov (ESRF), at beamline ID23-1. ©ESRF/C. Argoud |
References
Gushchin, I. et al., Science, 18 May 2017. DOI: 10.1126/science.aah6345.
Melnikov, I, et al., Science Advances , 12 May 2017. DOI: 10.1126/sciadv.1602952.



