- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2016
- Structural biology
- Mechanistic insights into the deubiquitinases A20 and Cezanne
Mechanistic insights into the deubiquitinases A20 and Cezanne
Ubiquitination is one of the most common posttranslational modifications of proteins. Deubiquitinating enzymes (DUBs) of the OTU family hydrolyse differently-linked polyubiquitin chains with high specificity. Crystal structures of OTU DUBs alone and trapped substrate and product complexes have revealed that the Lys11-linkage specificity of OTUD7B/Cezanne originates from large-scale conformational changes whereby ubiquitin-binding sites are formed and lost during a ubiquitin-assisted catalytic reaction.
Deubiquitinating enzymes (DUBs) are key regulators of many cellular processes. Their action can stabilise proteins from proteasomal degradation, or counteract ubiquitin-dependent signalling processes. Amongst the ~100 human DUBs are 16 OTU domain-containing enzymes with diverse cellular functions [1]. OTU DUBs can be further divided in subfamilies according to the size of their catalytic domain, which has been well studied for several enzymes. A20-like OTU DUBs feature the largest catalytic domain. The founding member A20 is an important negative-feedback regulator of NFκB signalling and a tumour suppressor in human cancers. Cezanne regulates NFκB as well as HIF1α signalling. A key difference between A20 and Cezanne is their ubiquitin chains specificity: while the A20 catalytic domain targets mostly Lys48-linked ubiquitin chains in vitro, Cezanne as well as the closely-related Cezanne2 are the only known DUBs with specificity for Lys11-linked polyubiquitin [2]. The molecular basis of this specificity has remained largely unclear.
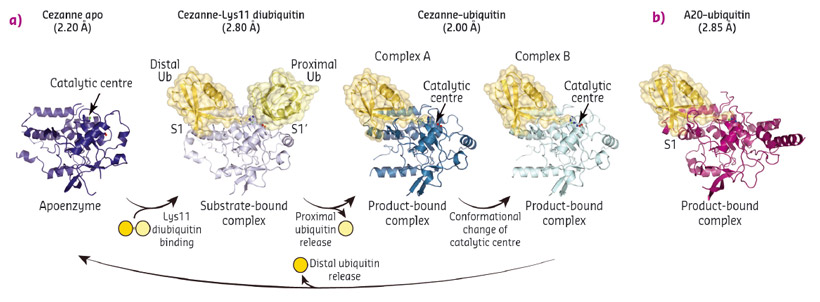
To understand linkage preference in A20-like OTU DUBs, and the Lys11-specificity of Cezanne in particular, we determined crystal structures of Cezanne alone, of Cezanne bound to Lys11-linked diubiquitin (substrate complex), and of Cezanne and A20 with monoubiquitin bound (product complexes) (Figure 92). The key to determining the complex structures were significant advances in chemical biology. The A20–ubiquitin complex structure exploited a new Cys-attacking propargyl warhead that allowed purification and crystallisation. Moreover, a Lys11-linked diubiquitin activity-based probe (ABP) was instrumental in the generation of a Cezanne–Lys11 diubiquitin complex. This probe covalently reacts with the catalytic Cys residue of Cezanne and enabled purification of the complex. Data for the Cezanne apo structure were collected at beamline ID23-1 and phased using SeMet phases derived from data collected at KEK. Data for the A20–ubiquitin structure was collected at beamline ID29, while the remaining Cezanne structures were collected at Diamond Light Source.
 |
|
Fig. 92: a) Crystal structures of Cezanne alone, bound to its substrate Lys11-linked diubiquitin and with the cleavage product monoubiquitin. b) Structure of monoubiquitin-bound A20. |
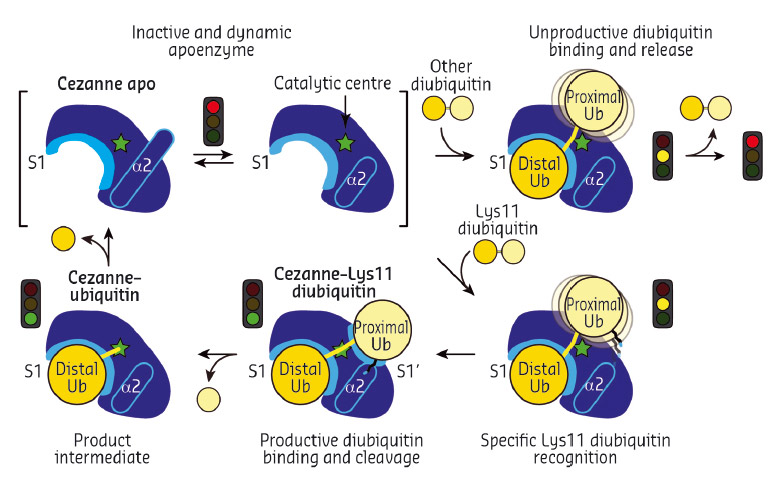
Collectively, the structures for the first time followed a DUB, Cezanne, along the reaction coordinate through the various stages of its catalytic cycle (Figure 92). Most striking, and so far only observed in Cezanne, were large-scale conformational changes which transformed an inactive to an active enzyme, and which modulated ubiquitin binding. Two ubiquitin binding sites are required to place a diubiquitin substrate across the active site; of these, the S1 ubiquitin-binding site that is occupied in monoubiquitin complexes, was present in all structures, and was identical between A20 and Cezanne. The second, so-called S1' ubiquitin-binding site, contacts the 'proximal' ubiquitin that determines the linkage type of the preferred substrate. Surprisingly, we found that in Cezanne, this binding site only exists in the diubiquitin complex structure, and is formed in situ when a substrate is bound, but lost in the apoenzyme and monoubiquitin-bound complex structures (Figure 93).
 |
|
Fig. 93: Model of the catalytic mechanism for the Lys11-specific DUB Cezanne. Cezanne recruits diubiquitin via the S1 site, yet only Lys11-chains can bind productively across the catalytic centre. |
The observed conformational changes were reconciled by hydrogen-deuterium exchange mass spectrometry (HDX-MS) experiments, and by mutagenesis and kinetic analysis. The latter further revealed a mechanism of substrate-assisted catalysis. Cleavage kinetics for differently-linked chain types showed similar KM values, but differed in kcat, with Lys11-linked diubiquitin cleavage being > 270-fold faster than other chain types. The diubiquitin complex structure revealed points of contact between the proximal ubiquitin and the enzyme that when interrupted led to a decreased kcat. After the OTU enzyme OTULIN, this is the second time such mechanism of ubiquitin-assisted catalysis has been identified to determine chain linkage specificity [3].
DUBs are emerging as promising drug targets in numerous human diseases, including cancer and neurodegeneration. Our work has revealed how new conformational states can occur while the enzymes perform their tasks. Knowledge of these states will be essential for rational drug design, and may enable new strategies to target DUBs with specific small molecule inhibitors.
Principal publication and authors
Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne, T.E.T. Mevissen (a), Y. Kulathu (a), M.P.C. Mulder (b), P.P. Geurink (b), S.L. Maslen (a), M. Gersch (a), P.R. Elliott (a), J.E. Burke (a), B.D.M. van Tol (b), M. Akutsu (a), F. El Oualid (b), M. Kawasaki (c), S.M.V. Freund (a), H. Ovaa (b) and D. Komander (a), Nature 538, 402-405 (2016). doi: 10.1038/nature19836.
(a) MRC Laboratory of Molecular Biology, Cambridge (UK)
(b) Netherlands Cancer Institute (NKI), Amsterdam (The Netherlands)
(c) High Energy Accelerator Research Organization (KEK), Tsukuba (Japan)
References
[1] Komander et al., Nat Rev Mol Cell Biol 10, 550-63 (2009).
[2] Mevissen et al., Cell 154, 169-84 (2013).
[3] Keusekotten et al., Cell 153, 1312-26 (2013).



