- Home
- News
- Spotlight on Science
- Crystal structure...
Crystal structure unveils DNA protection mechanism
13-10-2023
Scientists have used beamline ID30A-1 to determine the crystal structure of a SGO1-cohesin protein complex involved in binding duplicated chromosomes together until they are ready to separate during cell division. This protection mechanism ensures the accurate distribution of genetic information to daughter cells.
Our DNA is propagated each time our cells divide. Abnormal cell division, where the genome is not evenly distributed, can lead to infertility, cancer or genetic disorders. Understanding the molecular mechanisms underlying successful cell division will therefore open up new opportunities for drug design and pharmacological targeting.
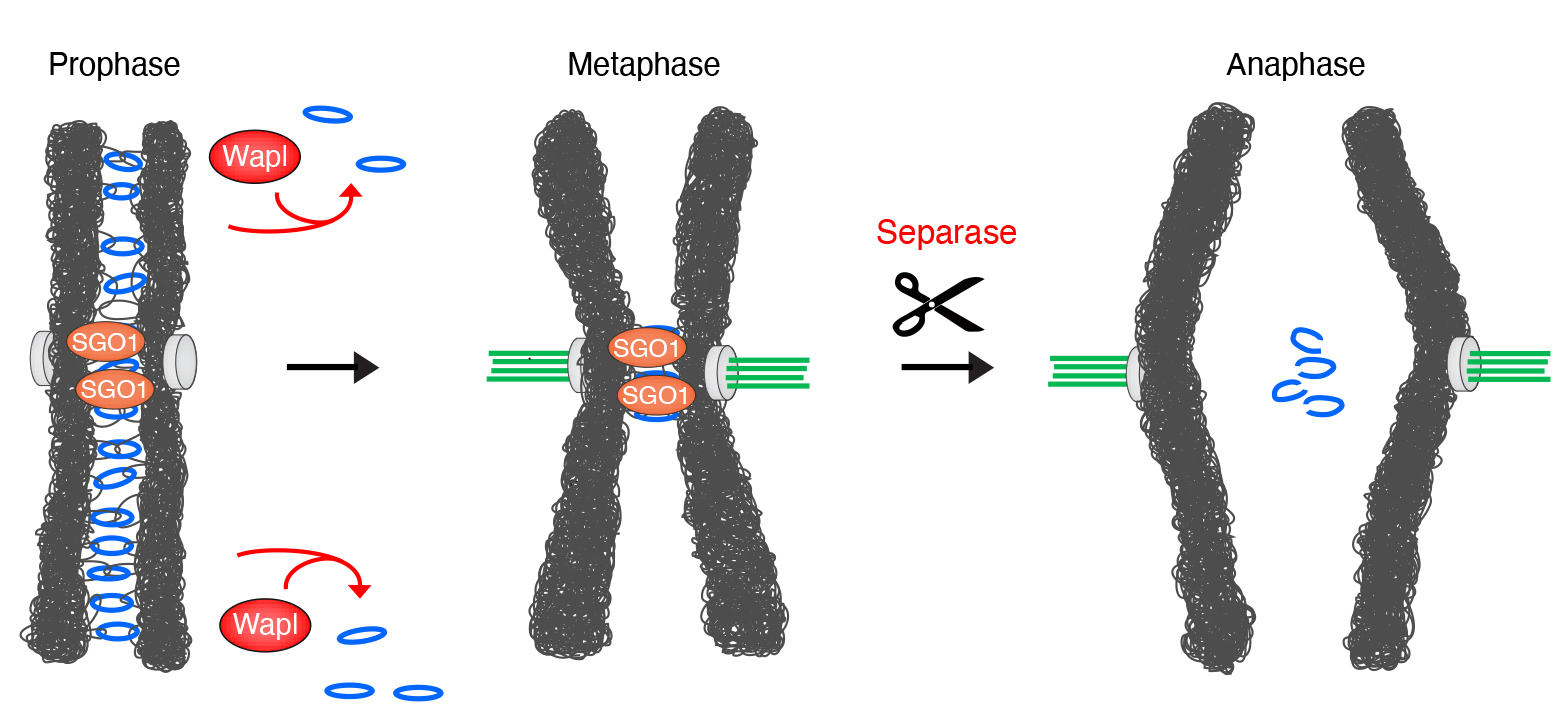
One key factor in this process is a protein complex called cohesin. Before cells divide, each chromosome is duplicated in the parent cell, with both copies bound together by cohesin proteins. Microtubules then project from opposite poles of the parent cell, attach to the centromeres of each chromosome, and align the chromosomes centrally within the cell, before pulling apart the two identical halves of each chromosome to an opposite pole of the cell. When the cell then splits in two, this yields two daughter cells with identical genetic content. In this process, cohesin is first removed from the arms of chromosomes by the cohesin release factor WAPL, but it is not removed from the centromere until each chromatid is correctly aligned and attached to the microtubule spindle apparatus (Figure 1). Cohesin removal from chromosome arms but not centromeres results in X-shaped chromosomes, one of the iconic images in biology. Cohesin protection at the centromeres involves a protein called SGO1, but the underlying mechanism was poorly understood.
Click image to enlarge
Fig. 1: Cohesin rings (blue) stably co-entrap the sister chromatids until the start of mitosis. Then, cohesin is removed from chromosome arms by the release factor WAPL. Centromeres are protected by SGO1 (Shugoshin). Centromeric cohesion is maintained until it is destroyed by Separase, which removes cohesin by proteolytically cleaving its SCC1 subunit at the metaphase-to-anaphase transition. This destruction is the trigger for chromosome segregation.
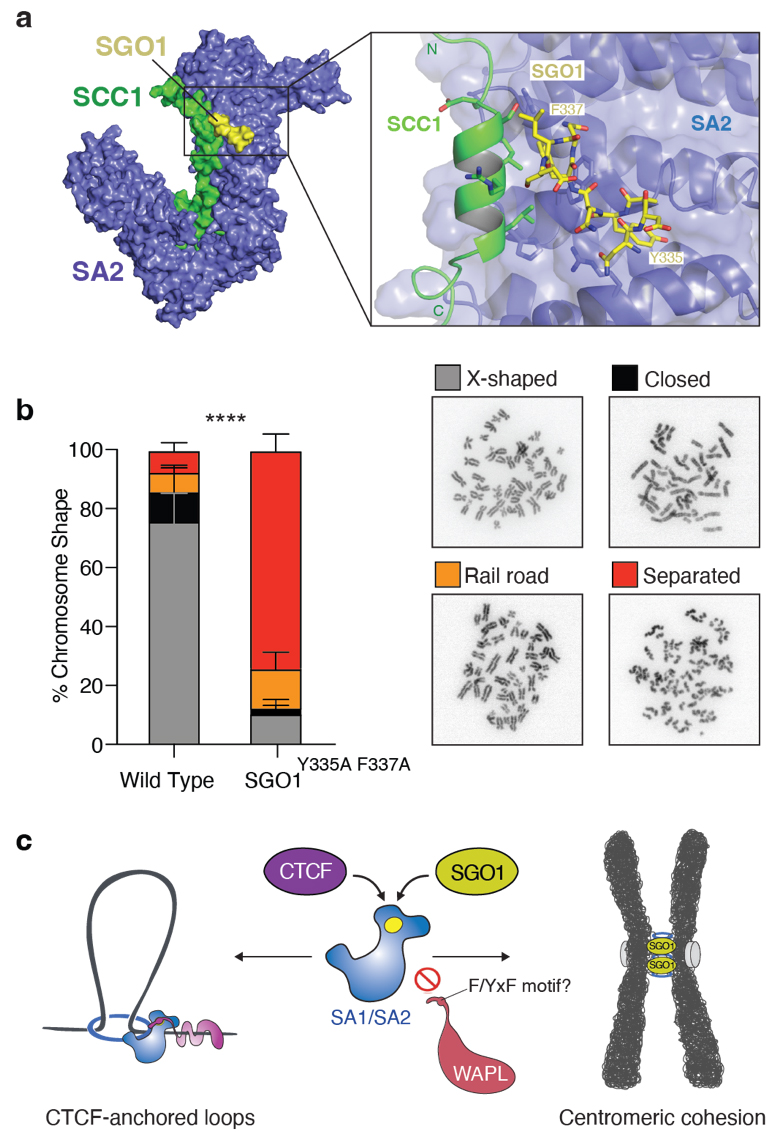
Cohesin is a 50-nm, ring-shaped complex comprising four core subunits: SMC1, SMC3, SCC1RAD21 and SA2, as well as several additional regulatory factors. Conserved amino acid segments from SGO1 interact with the cohesin subunits SA2 and SCC1RAD21. In this work, a SA2-SCC1RAD21-SGO1 complex was crystallised and its crystal structure determined to 3.2 Å resolution using data collected at the high-throughput beamline ID30A-1/MASSIF-1 [1]. The structure revealed that SGO1 binds to a conserved surface at the interface of the SA2 and SCC1 subunits of cohesin (Figure 2a). Binding is mediated by a conserved YxF amino acid motif of SGO1.
Click image to enlarge
Fig. 2: a) X-ray crystal structure of the SA2-SCC1-SGO1 complex: SA2 (blue), SCC1 (green) and SGO1 (yellow). b) Left panel: Quantification of chromosome phenotypes in prometaphase wild-type and SGO1Y335A F337A cells (****P < 0.0001). Right panel: Images of different chromosome phenotypes during prometaphase. c) Cohesin is controlled by different chromosomal regulators through a shared mechanism: CTCF uses its YxF motif to bind cohesin’s SA2/SA1 subunits to stabilise CTCF-anchored DNA loops (left), whereas SGO1 uses its YxF motif to bind cohesin’s SA2/SA1 subunits to protect centromeric sister chromatid cohesion (right).
Structural and biochemical data revealed that key amino acid residues of the YxF motif are required for the interaction of SGO1 with SA2-SCC1RAD21. Mutation of these amino acids in cells showed that this interaction is essential for the proper localisation of SGO1 and for the protection of cohesin at centromeres (Figure 2b). SGO1 prevents cohesin removal at centromeres by direct competition with WAPL for binding to the SA2-SCC1RAD21 complex, thereby enabling centromeric cohesion, which ensures correct chromosome segregation.
The interactions captured in the cohesin-SGO1 structure are similar to those seen between cohesin and an architectural protein factor called CTCF. Although SGO1 and CTCF appear to bind to cohesin in very similar manners, they control very different chromosomal processes – previous work shows that CTCF binding to cohesin is required for the formation of DNA loops [2]. It thus appears that cohesin complexes are controlled through a universal mechanism, irrespective of whether these complexes build DNA loops or hold together sister DNAs at the centromere (Figure 2c). CTCF and SGO1 may be merely the tip of the iceberg: cohesin is involved in a range of additional chromosomal processes that could be regulated by similar cohesin ligands that all interfere with the WAPL release mechanism.
Principal publication and authors
Structural basis of centromeric cohesion protection, A. García-Nieto (a), A. Patel (b), Y. Li (c), R. Oldenkamp (a), L. Feletto (b), J.J. Graham (b), L. Willems (a), K.W. Muir (d), D. Panne (b), B.D. Rowland (a), Nat. Struct. Mol. Biol. 30, 853-859 (2023); https://doi.org/10.1038/s41594-023-00968-y
(a) Division of Cell Biology, The Netherlands Cancer Institute, Amsterdam (The Netherlands)
(b) Leicester Institute of Structural and Chemical Biology, Department of Molecular and Cell Biology, University of Leicester (UK)
(c) Department of Fundamental Microbiology, Faculty of Biology and Medicine, University of Lausanne (Switzerland)
(d) MRC Laboratory of Molecular Biology, Cambridge (UK)
References
[1] M.W. Bowler et al., J. Synchrotron Radiat. 22(6), 1540 (2015).
[2] Y. Li et al., Nature 578, 472 (2020).
| About the beamline: ID30A-1 / MASSIF-1 |
| ID30A-1 / MASSIF-1 is a unique facility for the high-throughput, fully automatic characterisation and data collection of crystals of macromolecules.
The service does the hard work of screening crystals or collecting data sets through the night, freeing researchers to spend time on more challenging data collection problems and study the underlying biology. Beam time is booked flexibly and samples then enter a queuing system. Users interact with the beamline by describing experimental requirements, which are used by the beamline software to set data collection parameters in a database, ISPyB, where results are also viewed and downloaded. The service is made possible using the latest automation, a highly intense X-ray beam (5 x 1012 ph/sec in a flexible spot between 100 and 10 µm diameter) and complex workflows that fully evaluate samples, centre the best volumes and collect diffraction data sets optimised for maximum resolution with minimised radiation damage. |