- Home
- News
- Spotlight on Science
- X-ray crystallography...
X-ray crystallography reveals a new link between metabolism and epigenetics
02-11-2023
Scientists have used beamline ID30A-1 to reveal how a metabolic enzyme best known for balancing nucleotides can also play a key role in the epigenetic regulation of fatty acid synthesis – a process important in type 2 diabetes and other diseases.
De novo lipogenesis (DNL) is the metabolic pathway used primarily by the liver and adipose (fat-storing) tissue to synthesise fatty acids from excess carbohydrates or other precursors [1]. An increased rate of DNL is observed in cancer and in many characteristically "Western" diseases of affluence, such as metabolic disorder, type 2 diabetes, chronic liver disease and cardiovascular disease [2]. In response to metabolic changes, the liver and adipose tissue regulate the expression of genes encoding proteins important for de novo lipogenesis (DNL) through a variety of mechanisms. One of these processes involves the introduction of acetyl groups into histones, the proteins that package DNA, without altering their DNA sequence, thereby affecting gene expression. However, the molecular details that underlie this level of regulation are poorly understood.
In this work, an unexpected breakthrough has come from an initially unrelated investigation into the hyper-acetylation of chromatin during mammalian sperm development. Among the most highly abundant factors in male germ cells that bind acetyl-coenzyme A (acetyl-CoA), a key metabolite for histone acetylation, the protein NME1 and its closely related variant NME2 were identified. These ubiquitous metabolic enzymes, collectively called NME1/2, are best known for balancing proportions of nucleotides in the cell. They perform this role by transferring a phosphate group from a nucleoside triphosphate, typically ATP, to a nucleoside diphosphate. The surprising discovery of NME1/2 as a factor that also binds to acetyl-CoA prompted a more detailed investigation via a combination of structural, biophysical, genetic and epigenetic approaches.
Click image to enlarge
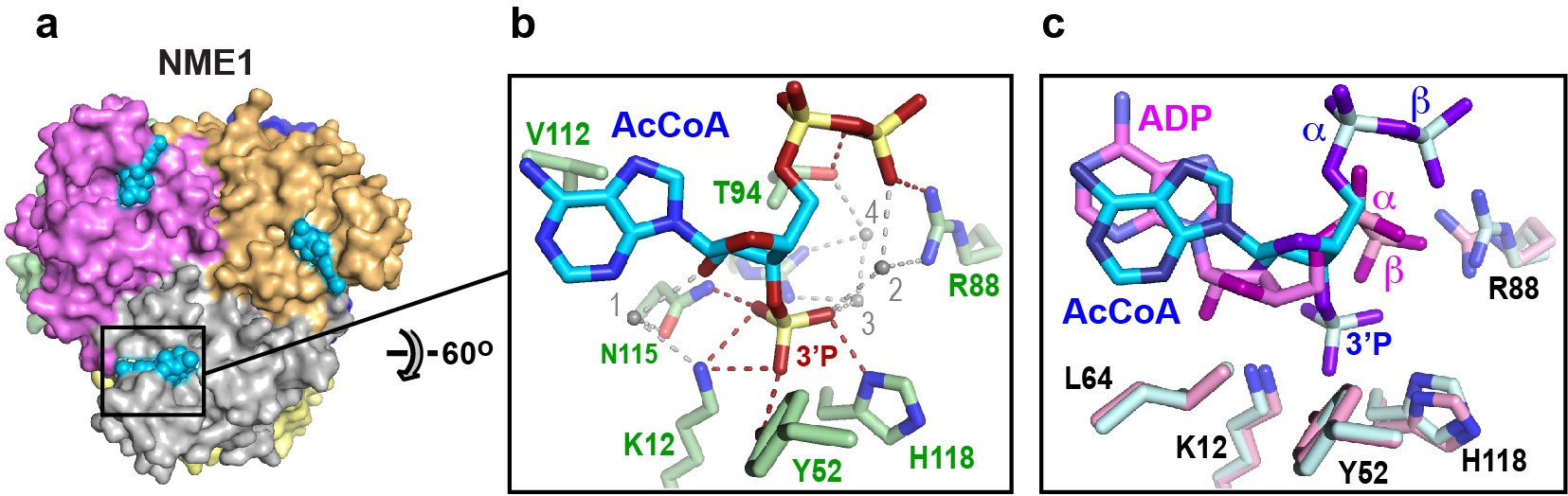
Fig. 1: Ligand recognition by NME1. a) Crystal structure of NME1, a hexameric enzyme with D3 symmetry. b) Details of the recognition of the nucleotide moiety of acetyl-CoA (AcCoA). Direct and water-mediated hydrogen bonds between NME1 and acetyl-CoA are shown as red and grey dashed lines, respectively. c) Superimposition of the acetyl-CoA and ADP ligands showing the distinct binding mode of acetyl-CoA, achieved through a rotation of the adenine base and a repositioning of the α- and β-phosphate groups.
Crystallographic data collected at beamline ID30A-1 revealed that NME1 recognises acetyl-CoA through the same binding site that recognises ATP and other nucleotides. Despite the similarity with ATP, NME1 recognises the nucleotide moiety of acetyl-CoA by a distinct binding mode that is critically dependent on the ligand's 3' phosphate group (Figure 1). Meanwhile, native mass spectrometry combined with biochemical assays showed that acetyl-CoA binding by NME1 is inhibited by ATP-mediated phosphorylation and is hence sensitive to the cellular energy status. Strikingly, when fed a high-fat diet, a mouse knockout model with greatly reduced NME1/2 levels exhibited excessive triglyceride synthesis and liver steatosis (fat buildup). In liver cells, NME1/2 was found to mediate a gene transcriptional response to a high-fat diet via the targeted acetylation of histones that inhibits fatty acid accumulation by repressing genes encoding key transcription factors involved in DNL and fatty acid metabolism.
These findings identify NME1/2 as a key regulator of the competing processes of histone acetylation and fatty acid synthesis. The results shed important light on how DNL is regulated and reveal a previously unsuspected level of cross-talk between metabolic and epigenetic pathways.
Principal publication and authors
Nucleoside Diphosphate Kinases 1 and 2 regulate a liver protective response to a high-fat diet, D. Iuso (a), I. Garcia-Saez (b), Y. Couté (c), Y. Yamaryo-Botté (a), E. Boeri Erba (b), A. Adrait (c), N. Zeaiter (d), M. Tokarska-Schlattner (d), Z. Macek Jilkova (a,e), F. Boussouar (a), S. Barral (a), L. Signor (b), K. Couturier (d), A. Hajmirza (a), F. Chuffart (a), E. Bourova-Flin (a), A.-L. Vitte (a), L. Bargier (f), D. Puthier (f), T. Decaens (a,e), S. Rousseaux (a), C. Botté (a), U. Schlattner (g), C. Petosa (b), S. Khochbin (a), Sci. Adv. 9, eadh0140 (2023); https://doi.org/10.1126/sciadv.adh0140
(a) Univ. Grenoble Alpes, CNRS, INSERM, Institute for Advanced Biosciences, La Tronche (France)
(b) Univ. Grenoble Alpes, CNRS, CEA, Institut de Biologie Structurale, Grenoble (France)
(c) Univ. Grenoble Alpes, INSERM, CEA, BioSanté, Grenoble (France)
(d) Univ. Grenoble Alpes, INSERM, Laboratory of Fundamental and Applied Bioenergetics, Grenoble (France)
(e) CHU Grenoble Alpes, Service d’hépato‐gastroentérologie, La Tronche (France)
(f) Aix Marseille Université, INSERM, TAGC, TGML, Marseille (France)
(g) Univ. Grenoble Alpes, INSERM, Institut Universitaire de France, Laboratory of Fundamental and Applied Bioenergetics, Grenoble (France)
References
[1] M. Wallace, C.M. Metallo, Semin. Cell Dev. Biol. 108, 65-71 (2020).
[2] F. Ameer et al., Metabolism 63, 895-902 (2014).
| About the beamline: ID30A-1 / MASSIF-1 |
|
ID30A-1/MASSIF-1 is a unique facility for high-throughput, fully automated characterisation and data collection of macromolecular crystals. The service automates the laborious tasks of crystal screening and data collection overnight, enabling researchers to focus on more challenging data analysis and biological studies. Beamtime is booked flexibly, and samples are then entered into a queuing system. Users interact with the beamline by describing their experimental requirements, which are used by the beamline software to set data collection parameters in a database called ISPyB. Results are also viewed and downloaded from ISPyB. The service is made possible using the latest automation, a highly intense X-ray beam (5 x 1012 ph/sec in a flexible spot between 100 and 10 µm diameter) and complex workflows that fully evaluate samples, centre the best volumes and collect diffraction data sets optimised for maximum resolution with minimised radiation damage. |