- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2006
- Structural Biology
- Crystal structure of human cytochrome P450 2D6
Crystal structure of human cytochrome P450 2D6
Cytochrome P450 2D6 is a haem-containing enzyme which catalyses the metabolism of at least 20% of known drugs. It exhibits considerable genetic polymorphism, affecting the rates of metabolism of some important drug molecules. Substrate metabolism is achieved by activation of molecular oxygen by the haem group in a process involving the delivery of two electrons to the P450 system followed by cleavage of the dioxygen bond to give an activated iron-oxygen species. This can then react with substrates. 2D6 typically recognises substrates containing a basic nitrogen and an aromatic ring - features which are especially found in a large number of central nervous system and cardiovascular drugs.
We determined the crystal structure of 2D6 to a resolution of 3.0 Å using data collected on ESRF beamline ID23-1. Crystallisation was carried out using a novel free interface diffusion technique, leading to rectangular plates with maximum dimensions of about 80 x 20 x 10 µm. The crystals suffered from considerable radiation damage during data collection and it was necessary to build up a complete dataset using data from multiple crystals. The structure was solved by molecular replacement, with the crystal asymmetric unit containing four 2D6 molecules, related by 222 symmetry.
 |
|
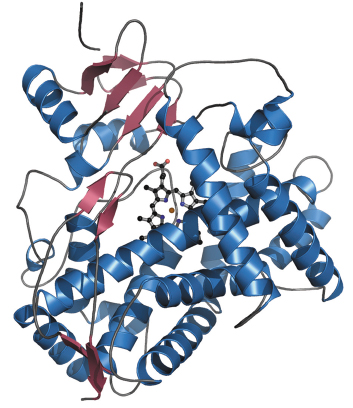
Fig. 75: A ribbon diagram of the 2D6 structure. |
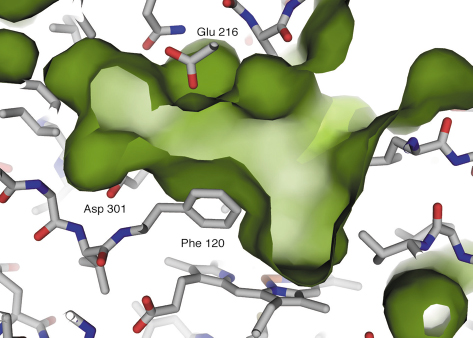
The 2D6 molecule has the characteristic P450 family fold (Figure 75), with the lengths and orientations of the individual secondary structural elements being very similar to those of the related 2C9 isozyme. There are, however, six main areas where large differences exist, several of which are directly involved in defining the shape and character of the 2D6 active site. There is a well-defined cavity above the haem group (Figure 76), bordered by many important residues previously implicated in substrate recognition and binding. The structure helps to explain how two of these residues, Asp301 and Glu216, can act as substrate binding residues and suggests that the role of Phe120 is to control the orientation of the substrate’s aromatic ring above the haem. The structure has also allowed an explanation of much of the published site-directed mutagenesis data and has helped in understanding the metabolism of several compounds, including the antihypertensive drug debrisoquine. Future structures of 2D6 with bound substrates and inhibitors will clarify the details of ligand recognition, and provide a structural basis for studying the effects of different 2D6 polymorphs on substrate metabolism.
 |
|
Fig. 76: The active site cavity above the haem group. Some key residues are labelled. |
Principal Publication and Authors
P. Rowland, F.E. Blaney, M.G. Smyth, J.J. Jones, V.R. Leydon, A.K. Oxbrow, C.J. Lewis, M.G. Tennant, S. Modi, D.S. Eggleston, R.J. Chenery and A.M. Bridges, J. Biol. Chem. 281, 7614-7622 (2006).
Molecular Discovery Research Department, GlaxoSmithKline (UK)



