- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2010
- Structural biology
- Activating the AP2 clathrin adaptor complex
Activating the AP2 clathrin adaptor complex
Compartmentalisation is a defining characteristic of eukaryotic cells that allows recognition events and chemical reactions to take place in distinct sub-cellular environments that are topographically equivalent to the extracellular space but within a cell's external boundary. All signals, nutrients and pathogens that require access to the cell’s cytoplasm must cross a membrane and all contact with other cells and components of the blood and immune system must occur on a membrane’s extracellular side. A membrane's functions and consequently its identity is largely defined by the transmembrane proteins embedded in it. The localisation and movement of transmembrane proteins between the cell’s various membranes is carried out by the cell's vesicular transport system.
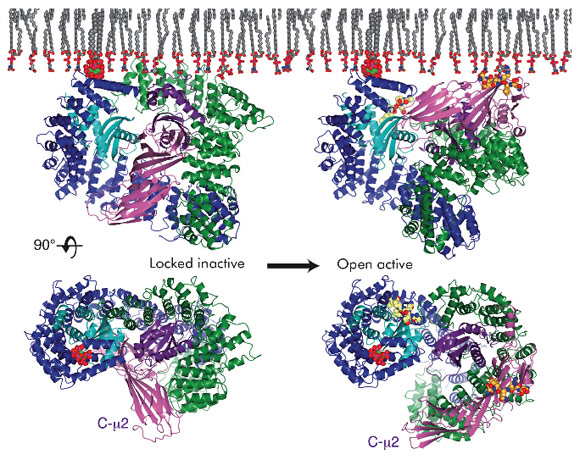
Coated transport vesicle formation requires the complex interplay of many proteins that are recruited from the cytoplasm, the membrane itself and the many types of transmembrane protein cargo that need to be selected for inclusion into the vesicle. Cargo is typically selected by the binding of short, linear amino acid motifs within the cytosolic portions of cargo to components of vesicle coats. Clathrin-coated vesicles (CCVs) mediate many post-Golgi trafficking routes including internalisation from the plasma membrane, termed clathrin-mediated endocytosis, and can be considered as having a 3-layered structure. The AP2 adaptor complex is a heterotetramer (subunits named α/β2/µ2/σ2) and is the most abundant endocytic clathrin adaptor [1]. It crosslinks the outer polymeric clathrin scaffold of endocytic CCVs to their inner membrane layer by binding directly both to the membrane phospholipid PtdIns4,5P2 and to cargo containing YxxΦ or ExxxLL motifs. In the 'locked' cytosolic form of the AP2 adaptor complex, whose crystal structure was determined from data collected at ESRF in 2002 [2], the binding sites for both YxxΦ motifs (on the C-terminal domain of the µ2 subunit, C-µ2) and [ED]xxxL[LI] motifs (on the σ2 subunit) are blocked by parts of β2 subunit. Thus, away from the membrane AP2 is unable to bind to either of its cognate cargo motifs.
The crystal structure of the open, motif-liganded form of the AP2 adaptor complex was recently determined from data collected at beamline ID29. Here, when compared to the structure of the locked form, C-µ2 undergoes a rotation of 130° and a translation of 40 Å to become relocated to an orthogonal face of the complex (Figure 108). The previously unstructured µ2 linker that connects C-µ2 to the rest of the complex becomes helical and binds back onto the complex. Four PtdIns4,5P2 and two endocytic motif-binding sites on AP2 also become co-planar, facilitating their simultaneous interaction with a PtdIns4,5P2/cargo-containing membrane. The conformational changes observed result in the unblocking of both motif-binding sites allowing them to bind cargo, which can freely diffuse into their binding sites through the plane of the membrane. Polarisation anisotropy measurements show that 99.9% of AP2 exists as the locked inactive form in the cytoplasm and that the structural rearrangements that facilitate cargo binding are driven by the association of AP2 with PtdIns4,5P2-containing membranes. It therefore appears that AP2 functions as a plasma membrane-activated switch for endocytic cargo recognition. This ensures that AP2 binds only to cargo when YxxΦ or ExxxLL motifs are displayed in the correct membrane, and that AP2 does not bind inappropriately to these rather non-specific short amino acid sequences elsewhere in the cell.
Principal publication and authors
L.P. Jackson (a), B.T. Kelly (a), A.J. McCoy (a), T. Gaffry (b), L.C. James (c), B.M. Collins (d), S. Höning (c), P.R. Evans (b) and D.J. Owen (a), Cell 141, 1220-1229 (2010).
(a) Cambridge Institute for Medical Research, University of Cambridge (UK)
(b) Institute of Biochemistry I, University of Cologne (Germany)
(c) Medical Research Council Laboratory of Molecular Biology, Cambridge (UK)
(d) Institute for Molecular Bioscience, The University of Queensland, Brisbane (Australia)
References
[1] M.S. Robinson, J Cell Biol. 104, 887-95 (1987).
[2] B.M. Collins, A.J. McCoy, H.M. Kent, P.R. Evans and D.J. Owen, Cell 109, 523-535 (2002).