- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2013
- Structural biology
- Structural basis for ion permeation in a pentameric ligand-gated ion channel revealed by X-ray crystallography
Structural basis for ion permeation in a pentameric ligand-gated ion channel revealed by X-ray crystallography
Pentameric ligand-gated ion channels (pLGICs) are a major family of membrane receptors that mediate fast chemical transmission of nerve signals in the central and peripheral nervous system [1]. In vertebrates, the family encompasses the cationic selective acetylcholine and serotonine receptors, and the anionic selective glycine and γ-amino-butyric acid receptors. pLGICs are dynamic proteins that couple neurotransmitter binding in their extracellular-domain (ECD) to the opening of ion channels embedded in their transmembrane domain (TMD). The receptors share a common architecture with a large ECD and a TMD that is composed of four α-helices, named M1 to M4, that form a transmembrane pore bordered by the M2 helices (Figure 52a). The flow of ions across the membrane is inhibited by a structurally-diverse class of molecules including tricyclic antidepressants, local anaesthetics and certain transition metals that bind to the transmembrane pore. The molecular understanding of ion permeation is thus a central issue in the study of these ion channels.
 |
|
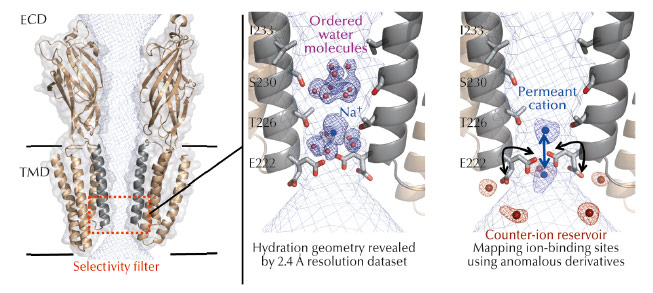
Fig. 52: Left: Cartoon representation of the GLIC ion channel viewed from the side (for clarity only two subunits are shown). The solvent-accessible surface of the channel is shown as a mesh (light blue). The TMD forms a narrow channel lined by the M2 helices from each subunit (grey). The selectivity filter is highlighted in red. Middle: Structurally ordered cation and pentamers of electron density and water molecules in the pore of GLIC. The blue mesh shown is the 2mFo-DFc electrodensity map. Right: Cation and anion binding sites revealed by the use of anomalous scattering from derivatives. The anomalous difference maps calculated for Br–, Cs+ or Rb+ are superimposed and shown as meshes. |
The crystal structure of GLIC, a prokaryotic homolog of the family was solved in an open form of the channel and provided a significant advance in the possibility to study ion permeation [2-3]. The pore of GLIC is funnel-shaped, with a constriction of 5 Å diameter at the Thr226 and Glu222 level that hosts the charge selectivity filter (Figure 52a). However, the molecular mechanisms of ion permeation in pLGICs are complex mechanisms involving protein residues, ions and water molecules that interact together dynamically and transiently as the ions flow down the channel. A proper description of these is difficult to derive from existing structures given the limited resolution achieved.
To understand the molecular mechanisms of ion permeation, we solved the crystal structure of GLIC at 2.4 Å, revealing for the first time the hydration geometry in the pore of a pLGIC (Figure 52b). The crystal structure reveals ordered water molecules at the level of two rings of hydroxylated residues (named Ser230 and Thr226). Here, two water molecule pentagons are observed sandwiching a sodium ion between them. Simulations that pull a cation through the pore reveal that the water pentagons observed in the crystal structure actively contribute to cation translocation. More precisely, these ordered water molecules contribute to lower the energy barriers encountered by the permeant ion when it crosses the hydrophobic constriction barriers that are located along the selectivity filter.
To better understand the function of the organised water molecules observed in the pore, it was necessary to identify the monovalent ion binding sites in the transmembrane pore. With this aim, the protein was co-crystallised with Br-, Cs+, or Rb+ and X-ray data were collected at optimised wavelengths for each of these derivatives so as to maximise the anomalous signal (Figure 52c). Some of these experiments were performed at beamline ID23-1, taking advantage of the tuneable wavelength of the beamline. The permeating ions occupy two preferential positions in the most constricted region of the pore that hosts the selectivity filter. In the GLIC structure, the Glu222 carboxylate groups are flexible and continuously exchange between alternative conformations, thus facilitating ion transport. Finally, the presence of a reservoir of counter-ions located at the intracellular mouth of the pore might help in either retaining the permeant ion or creating a favourable corridor through it.
 |
|
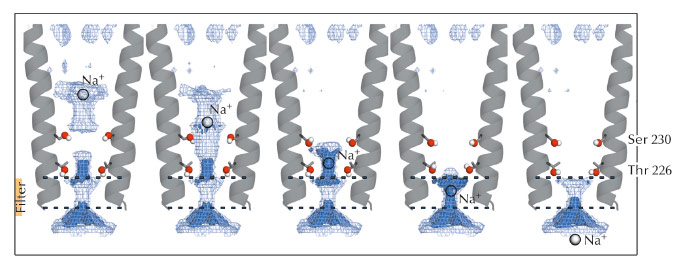
Fig. 53: Optimised set of configurations of the Ser230 and Thr226 side chains can maximise the permeant ion density at five levels in the pore, from its extracellular to its intracellular end. |
Electrostatic calculations illustrated the roles of residues Ser230 and Thr226 during permeation. Both residues were shown to facilitate ion transport by reducing electrostatic free energy barriers encountered by the ion during its translocation (Figure 53). Due to the strong sequence conservation of M2 pore-lining residues within the pLGIC family, the conclusions presented in this study might be transferable to all the pLGIC channels.
Principal publication and authors
L. Sauguet (a,b), F. Poitevin (a), S. Murail (c), C. Van Renterghem (b), G. Moraga-Cid (b), L. Malherbe (a,b), A.W. Thompson (d), P. Koehl (e), P.J. Corringer (b), M. Baaden (c) and M. Delarue (a), EMBO journal 32, 728-41 (2013).
(a) Unité de Dynamique Structurale des Macromolécules, Institut Pasteur, CNRS UMR3528, Paris (France)
(b) G5 Récepteurs-Canaux, Institut Pasteur, CNRS URA2182, Paris (France)
(c) Laboratoire de Biochimie Théorique, IBPC, CNRS UPR9080, Université Paris Diderot Sorbonne Paris Cité, Paris (France)
(d) Synchrotron Soleil, Saint Aubin (France) (e) Genome Center, Department of Computing science, UC Davis, CA (USA)
References
[1] P.J. Corringer et al, Structure 20, 941-56 (2012).
[2] N. Bocquet et al, Nature 457, 111-114 (2009).
[3] R. Hilf and R. Dutzler, Nature 457, 115-118 (2009).



