- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2013
- Structural biology
- How arrestin is activated for GPCR binding
How arrestin is activated for GPCR binding
The large family of G-protein-coupled receptors (GPCRs) controls diverse sensory and physiological responses and is thus a major focus of research and a central drug target. These receptors bind a variety of ligands (e.g. light, odorants, hormones, neurotransmitters) and communicate this signal into the cell by coupling to intracellular proteins [1]. One of these binding partners is arrestin, of which the canonical role is either to stop signalling by blocking receptor coupling to G-proteins, or to initiate arrestin-dependent signalling by bringing together other signalling proteins. We have used protein X-ray crystallography to gain insight into how arrestins are activated for receptor binding.
 |
|
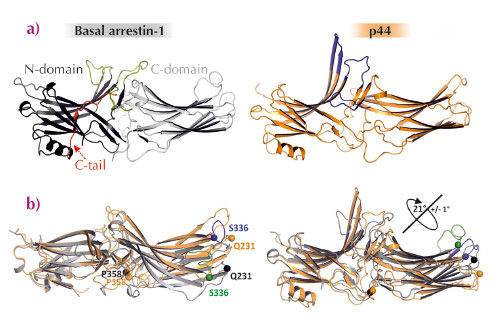
Fig. 57: Structural differences between basal arrestin-1 and p44. a) Ribbon diagrams of basal arrestin-1 (PDB entry 1CF1) and p44. The N- and C-domains of basal arrestin-1 are coloured dark-grey and light-grey, respectively, the C-tail is shown in red and important loops in the central crest region are coloured green. p44 is shown in orange, and key loops coloured blue. b) Side-views of the interdomain rotation in p44 (orange) as compared to basal arrestin-1 (grey). The N-domains are superimposed to show the rotation, evidenced by the change in position of the specific residues S336, Q231 as compared to the reference residue P358. |
Structurally, arrestins are composed of two near-symmetric lobes (N- and C-domain) that are stabilised by a long C-terminal tail (C-tail) (Figure 57a). In their basal state, arrestins are unable to recognise and bind an activated receptor, which must first be phosphorylated by a kinase. and then the receptor-attached phosphates displace the C-tail of arrestin thereby activating arrestin to bind the receptor. Although the crystal structure of basal arrestin was first solved some fifteen years ago, this structure did not indicate how C-tail displacement activates arrestin.
We tackled this problem using X-ray crystallography and site-directed fluorescence spectroscopy on a pre-activated form of arrestin-1, the arrestin subgroup present in the rod cells of the retina.
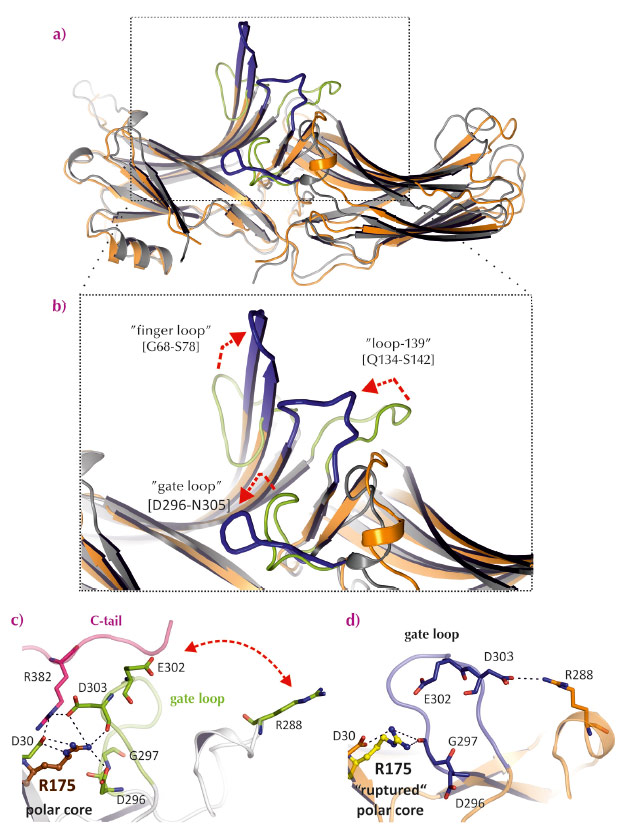
We pre-activated arrestin by removing its C-tail by mutation and crystallising it in the presence of the GPCR opsin, which was most likely activated by the crystallisation conditions [2,3]. The crystal structure of C-terminally truncated arrestin (called p44) was solved at 3.0 Å resolution using diffraction data collected at beamline ID14-4. It differs substantially with that of basal arrestin (Figure 57a). Several of the hydrogen-bond networks that stabilise the orientation of the two lobes in basal arrestin are broken in p44, which results in the two lobes of p44 being rotated 21° with respect to basal arrestin-1 (Figure 57b). Additionally, several of the flexible loops in the receptor-binding surface of arrestin are significantly rearranged in p44 (Figure 58). Overall, the structure of p44 indicates a general mechanism of arrestin activation, whereby C-tail displacement increases flexibility in critical regions of the arrestin molecule that allow a proper fitting to the active receptor.
 |
|
Fig. 58: Comparison of loops that differ between basal arrestin-1 and p44. a) and b) Overall structure and close-up view of superimposed structures of basal arrestin-1 and p44 (coloured as in Figure 57a). Critical loops involved in receptor binding are labelled with red arrows indicating activating changes in the structure of these loops. c) and d) The hydrogen-bond network linking the gate loop (green), the C-tail (red) and R175 in the central polar core region of basal arrestin-1 (grey) is broken in p44 (blue/orange). |
This activation mechanism is most likely conserved within the small arrestin family (four subgroubs) and this explains how hundreds of different GPCRs are bound by only a few arrestin variants. The overarching theme of activation is an increase in conformational flexibility, which presumably facilitates arrestin binding to different GPCRs at variable receptor stoichiometry [4] as well as to a multitude of other binding partners.
Our structure of p44 was published in the same journal issue as another report of pre-activated arrestin crystal structure by B.Kobilka and R.Lefkowitz, who shared the Nobel Prize for Chemistry in 2012. That study describes an arrestin that is fixed by a synthetic antibody fragment and bound to a phosphorylated peptide-mimic of the phosphorylated C-terminus of an active GPCR [5]. Remarkably, the two structures are so similar they validate one another and the conclusions concerning arrestin activation described above.
Principal publication and authors
Y.J. Kim (a), K.P. Hofmann (a), O.P. Ernst (b), P. Scheerer (c), H.W. Choe (a,d) and M.E. Sommer (a), Nature 497, 142-146 (2013).
(a) Institut für Medizinische Physik und Biophysik (IMPB), Charité-Universitätsmedizin Berlin (Germany)
(b) Departments of Biochemistry and Molecular Genetics, University of Toronto (Canada)
(c) IMPB, AG Protein X-ray Crystallography, Charité-Universitätsmedizin Berlin (Germany)
(d) Department of Chemistry, College of Natural Science, Chonbuk National University (South Korea)
References
[1] K.P.Hofmann et al., Trends Biochem Sci. 34, 540-552 (2009).
[2] P.Scheerer et al., Nature 455, 497-502 (2008).
[3] H.-W.Choe et al., Nature 471, 651-655 (2011).
[4] M.E.Sommer et al., Nat Comm 3, 995 (2012).
[5] A.K.Shukla et al., Nature 497, 137-141 (2013).



