- Home
- News
- Spotlight on Science
- High-pressure cryocooling...
High-pressure cryocooling of protein crystals reveals gas transport channels in a hydrogen-converting enzyme
28-06-2016
Networks of hydrophobic tunnels transport substrate and inhibitory gases (i.e. oxygen) to/from the active sites of NiFe hydrogenases. Here, the tunnel network of an oxygen-tolerant NiFe hydrogenase has been mapped by combining single-crystal X-ray diffraction with the derivatisation, under high pressure, of crystals with krypton gas. This mapping process revealed the presence of only two gas tunnels. It is postulated that the presence of so few channels allows for a much tighter control of the flow of gas molecules to the catalytic centre, thus explaining the remarkable oxygen tolerance of the enzyme studied.
NiFe hydrogenases are metalloenzymes that catalyse the splitting of molecular hydrogen into electrons and protons, which makes them potentially very interesting for developments in renewable energy technologies. While some NiFe hydrogenases are irreversibly inactivated in the presence of O2, others are tolerant to atmospheric levels of oxygen, making them easier to handle and thus more desirable targets for biotechnological applications.
Catalysis takes place at the NiFe active site, which is deeply buried within the protein and connected to the surface through hydrophobic gas tunnels. These tunnels are responsible for the transport of both substrate and inhibitory gases. Electrons are channelled from the active site via three FeS clusters to the primary electron acceptor, a cytochrome b562. However, pathways for the corresponding protons and the gas molecules (H2 and O2) were largely unknown.
Crystal structures of a NiFe hydrogenase that sustains catalytic activity even in the presence of oxygen [1, 2] revealed plausible pathways for electron transport and water release. To visualise the tunnel network, noble gas derivatisation was performed using a high-pressure cooling device recently developed at the ESRF [3]. The crystals were pressurised for 10 minutes under 80 bar of krypton gas, and then flash-cooled (while still under pressure), and finally depressurised and transferred in liquid nitrogen to beamline ID23-1 for the collection of X-ray diffraction data.
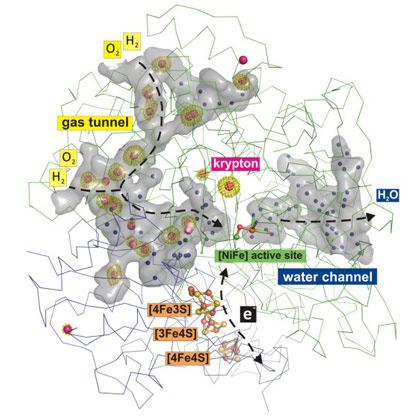
With this approach, 19 krypton sites could be identified inside the protein, which map out a hydrophobic gas tunnel network that connects the protein surface with the NiFe catalytic centre (Figure 1). On the other side of the protein, water molecules are found in a hydrophilic channel, which also links the active site with the protein surface (Figure 1).
 |
|
Figure 1. Gas and water tunnel networks in an O2-tolerant NiFe hydrogenase. Krypton atoms are shown as red spheres with green electron densities, and water molecules are shown as blue spheres. |
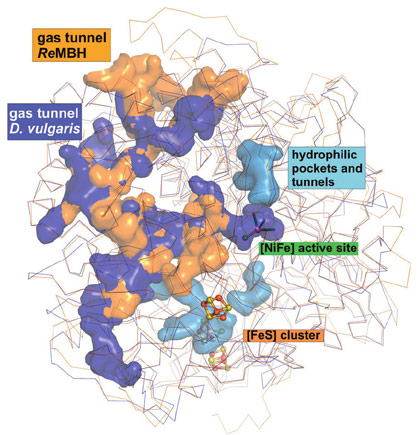
Concomitant calculations revealed considerable differences between O2-tolerant and O2-sensitive hydrogenases both in tunnel size and quantity, which might be related to their distinct susceptibility for O2. Notably, O2-sensitive NiFe hydrogenases have on average twice as many hydrophobic gas channels as the O2-tolerant ones (Figure 2). Thus, in these O2-tolerant hydrogenases, the existence of only two gas channels may allow for a much tighter control of the flow of gas molecules to the sensitive catalytic centre, therefore explaining their remarkable O2 tolerance.
 |
|
Figure 2. Comparison of the cavities in O2-tolerant (orange) and O2-sensitive (blue) NiFe hydrogenases. Hydrophilic water tunnels are shown in light blue. |
Principal publication and authors
Krypton derivatization of an O2-tolerant membrane-bound [NiFe] hydrogenase reveals a hydrophobic tunnel network for gas transport, J. Kalms (a), A. Schmidt (a), S. Frielingsdorf (b), P. van der Linden (c), D. von Stetten (c), O. Lenz (b), P. Carpentier (c), and P. Scheerer (a), Angew. Chem. Int. Ed. 55, 5586-5590 (2016); doi: 10.1002/anie.201508976.
(a) Institut für Medizinische Physik und Biophysik (CC2), Group Protein X-ray Crystallography and Signal Transduction, Charité – Universitätsmedizin Berlin (Germany)
(b) Institut für Chemie, Technische Universität Berlin (Germany)
(c) ESRF
References
[1] J. Fritsch, P. Scheerer, S. Frielingsdorf, S. Kroschinsky, B. Friedrich, O. Lenz, C. M. Spahn, Nature 479, 249–252 (2011).
[2] S. Frielingsdorf, J. Fritsch, A. Schmidt, M. Hammer, J. Löwenstein, E. Siebert, V. Pelmenschikov, T. Jaenicke, J. Kalms, Y. Rippers, F. Lendzian, I. Zebger, C. Teutloff, M. Kaupp, R. Bittl, P. Hildebrandt, B. Friedrich, O. Lenz, P. Scheerer, Nat. Chem. Biol. 10, 378–385 (2014).
[3] P. van der Linden, F. Dobias, H. Vitoux, U. Kapp, J. Jacobs, S. Mc Sweeney, C. Mueller-Dieckmann and P. Carpentier, J. Appl. Cryst. 47, 584-592 (2014).
Top image: Krypton atoms reveal the gas and water tunnel networks in an oxygen-tolerant NiFe hydrogenase.



