- Home
- News
- Spotlight on Science
- Four crystal structures...
Four crystal structures give insights into the enzymatic mechanism of transglutaminase 3
07-12-2023
Researchers have determined the crystal structures of a human enzyme in three different conformations, along with a patient autoantibody associated with dermatitis herpetiformis, a disease linked to gluten sensitivity. X-ray diffraction data collected at the ESRF show how calcium-ion and substrate binding dramatically changes the conformation of the enzyme. The results give insights into the disease mechanism of this autoimmune condition.
Dermatitis herpetiformis (DH) is a skin condition that manifests as skin blisters and itchy rashes in genetically susceptible individuals. Similar to celiac disease, it is caused by the ingestion of dietary gluten, prevalent in bread and other grain products. Both diseases have strong human leukocyte antigen (HLA) associations to HLA-DQ2/8, and they are both hallmarked by the formation of antibodies against human transglutaminases: transglutaminase 2 in celiac disease and transglutaminase 2 and 3 in DH. These enzymes are calcium-dependent and have the ability to bind and modify gluten peptides. The substrate-enzyme intermediate acts as an antigen, leading to release of deamidated gluten peptides in endosomes of transglutaminase 2- or transglutaminase 3- specific B-cells. Such B-cells will then present deamidated gluten peptides by the disease-associated HLA molecules to CD4+ T cells and receive help for autoantibody production.
Scientists at the University of Oslo have crystallised and solved four different transglutaminase 3 (TG3) structures in complex with a DH patient-derived antibody using macromolecular crystallography beamlines ID23-1, ID23-2, ID30A-3 and ID30B. TG3 has four domains: N-terminal, catalytic core and two C-terminal b-barrels: C1 and C2. To become catalytically active, the enzyme needs a proteolytical cleavage between the core domain and the C1C2 domain, along with the binding of three calcium ions.
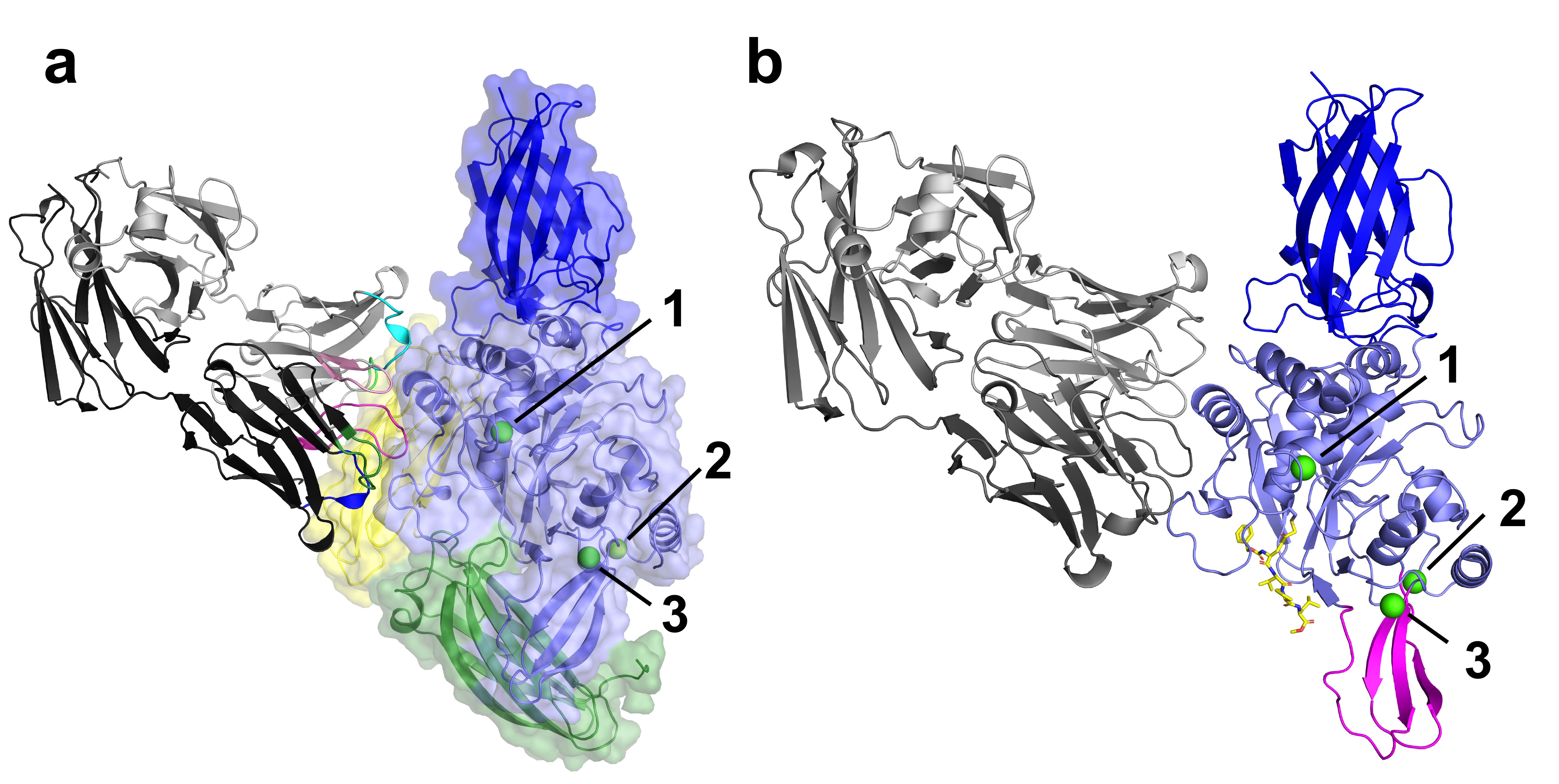
The structural analysis of four TG3 structures has provided valuable molecular insights into both TG3 enzymology and autoantibody binding. Notably, the structure without calcium ions shows a large conformational change, suggesting that binding of calcium leads to structural rearrangements important for the catalytic activity (Figure 1a). The native substrate of the enzyme, gluten peptides, binds by forming a thioester bond between a glutamine residue of the peptide and the active site cysteine-273 of TG3. This thioester intermediate can be resolved through a nucleophilic attack by a secondary substrate. In deamidation reactions, the secondary substrate is water, and the resulting product will be the conversion of glutamine to glutamate in the released gluten peptide. If the secondary substrate is a polyamine or the e-NH2 group of a polypeptide lysine residue, it results in transamidation. In this way, the enzyme creates an isopeptide bond between a glutamine side chain and a lysine side chain, forming a crosslink.
Click image to enlarge
Fig. 1: a) The crystal structure of TG3 (N-terminal domain in dark blue, catalytic domain in light blue, C1 domain in green and C2 domain in yellow) bound to autoantibody Fab DH63-B02 (shown in grey with coloured complementarity-determining loops) and calcium ions (green spheres, numbered). b) The crystal structure of TG3 (N-terminal domain in dark blue, catalytic domain in light blue, with moving b-sheet in magenta) bound to autoantibody Fab DH63-B02 (shown in grey), calcium ions (green spheres, numbered) and inhibitor Z-DON (yellow sticks). The C1 and C2 domains are missing from the structure.
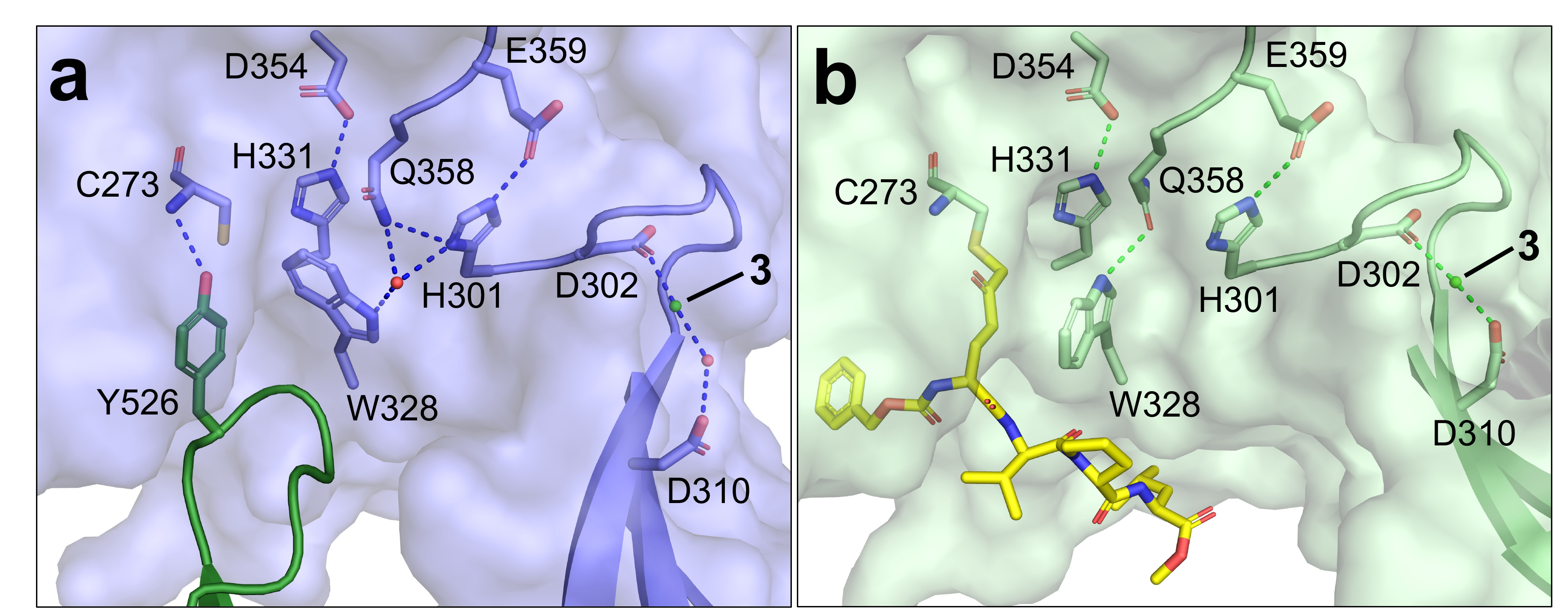
The structure with a gluten-substrate mimicking inhibitor (Z-DON) bound in the active site show a dramatic conformational change: two of the domains, C1 and C2, are missing and a b-sheet has moved to accommodate the inhibitor (Figure 1b). By using the inhibitor Z-DON, the enzyme is trapped in a substrate-bound intermediate through the formation of an irreversible alkylation at the active site. The details of the catalytic domain changes are shown in Figure 2. The hydrogen bond between the C1 domain residue tyrosine-526 and the catalytic domain residue cysteine-273 is broken, allowing Z-DON to covalently bind to cysteine-273, causing the nearby residue tryptophan-328 to rotate by 90 degrees. This rotation opens up a tunnel over the active site residues cysteine-273, histidine-331 and aspartate-354, preparing for the enzymatic reaction of transamidation. Taken together, the structures give valuable insight into the enzymatic mechanism of transglutaminase 3, as well as providing a model for autoantibody formation in DH.
Click image to enlarge
Fig. 2: Closeup of the active site of TG3. The catalytic triad of cysteine-273 (C273), histidine-331 (H331) and aspartate-354 (D354) is shown. Ca2+ is shown as a green sphere (label 3), a water molecule as a red sphere. H-bonds are indicated by dashed lines. a) TG3 (light blue), showing the C1 domain in dark green. b) TG3 (light green) with Z-DON (yellow sticks) bound.
Interestingly, it was observed that the two conformations of TG3 (with and without inhibitor) binds to autoantibodies with similar affinity. This further strengthens the hypothesis that B-cells specific for TG3 via involvement of the enzyme can capture gluten peptide substrates for presentation to pathogenic CD4+ T-cells.
The striking parallel between the role of TG3 in DH and TG2 in celiac disease highlights the importance of transglutaminases in autoimmune diseases. However, further investigation is needed to elucidate the active conformation of TG2 and the precise location of its conformational epitopes. Inspired by the findings of this study, future research should focus on a similar analysis of TG2 to further unravel its involvement in autoimmune disease.
Principal publication and authors
Autoantibody binding and unique conformation of human transglutaminase 3, J.E. Heggelund (a,b), S. Das (a,b), J. Stamnaes (a,b), R. Iversen (a,b), L.M. Sollid (a,b), Nat. Commun. 14, 6216 (2023); https://doi.org/10.1038/s41467-023-42004-z
(a) KG Jebsen Coeliac Disease Research Centre, Institute of Clinical Medicine, University of Oslo (Norway)
(b) Department of Immunology, Oslo University Hospital-Rikshospitalet, Oslo (Norway)
| About the beamlines |
| The ESRF’s Structural Biology group operates a world-leading suite of synchrotron radiation beamlines dedicated to the study of biological macromolecules, including two highly intense, tuneable beamlines (ID23-1 and ID30B), a unique beamline for fully automatic data collection (ID30A-1), two microfocus / minibeam beamlines dedicated to protein crystallography (ID23-2 and ID30A-3), a protein solution scattering beamline (BM29) and a cryo-EM microscope (CM01). For more information, please click on each beamline link. |