- Home
- News
- Spotlight on Science
- Snapshot of a bacterial...
Snapshot of a bacterial transporter in the act of secreting a protein
23-09-2011
The human urinary tract is one of the most common sites for bacterial infection and most of the diagnosed urinary tract infections are caused by the uropathogenic Escherichia coli. During the infection process, the bacteria recognise and adhere to host epithelial cells by using specific extracellular adhesive organelles termed pili. Despite the wealth of structural information, little is known about how pilus formation is orchestrated at the bacterial cell surface. Recently, a crystal structure has captured the pilus assembly platform in the act of secreting its cognate substrate. This breakthrough from the Waksman group (UK) and collaborators provides new insights into the molecular details of pilus assembly and will serve as a new basis for drug-design.
Share
The first stage of infection by pathogenic bacteria is the specific targeting of bacteria to the host tissues. This recognition and adherence event is mediated by fibres known as pili present at the bacterial cell surface. Because pili have no equivalent in eukaryotic organisms, they are very promising drug targets.
Uropathogenic Escherichia coli (UPEC) is the main causative agent of urinary tract infections, one of the most prevalent bacterial infections, afflicting mainly women. To attach to and invade the epithelium of the bladder, UPEC express the archetypal type 1 pili that are assembled by the so-called chaperone/usher (CU) pathway, one of the most characterised secretion systems among Gram-negative bacteria [1]. These pili are constituted by immunoglobulin(Ig)-like fold protein subunits that assemble at a defined assembly platform, the usher. The subunit first engaged in assembly is FimH, the adhesin at the very tip of the pilus. Once secreted in the periplasm through the general Sec machinery (which facilitates the translocation of proteins across and into biological membranes), each subunit is stabilised by a dedicated molecular chaperone, FimC, which prevents subunits from aggregating non-productively or polymerising prematurely. The chaperone-subunit complexes are then targeted differentially to the outer membrane usher protein, FimD, which catalyses assembly of the subunits into a linear fibre in a defined order.
Over the last two decades, some of the molecular details of pilus biogenesis through the CU pathway have been provided, including the elucidation of the structural basis of chaperone function and also the molecular characterisation of the subunits polymerisation process [1]. Subunits have a C-terminally truncated Ig-fold where strand G is missing entirely. The chaperone, in a process termed “donor-strand complementation”, donates one of its own strands to complement the subunit’s fold. Subunit polymerisation occurs in a mechanism termed “donor-strand exchange” whereby the chaperone strand is “exchanged” by the N-terminal extension of the subunit coming next in assembly.
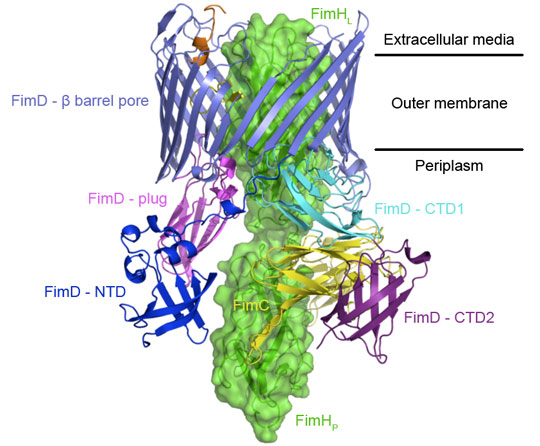
However, little is known about how the usher works. Here we report on a new crystal structure describing the first step in pilus biogenesis, i.e. the FimD usher bound to its cognate FimC:FimH substrate, using diffraction data collected at beamline ID29. This membrane protein complex of 150 kDa shows for the first time an export nanomachine caught in the act of secreting a protein substrate (Figure 1). The detailed structure solved at 2.8 Å resolution captures the full-length usher FimD in its active state, along with the apo state of the usher 24-stranded β-barrel pore domain [2], thereby elucidating the mechanism of pore activation.
Previous studies have shown that the chaperone-subunit complexes are specifically recruited by the N-terminal domain (NTD) of the usher [3]. The new FimD:FimC:FimH crystal structure reveals another binding site, the C-terminal domains (CTD1 and CTD2), which stabilise the adhesin subunit FimH still guarded by the chaperone but lined up inside the pore domain of the usher. A dramatic displacement of the plug domain from the lumen of the pore to the periplasmic space paves the way for subunit insertion.
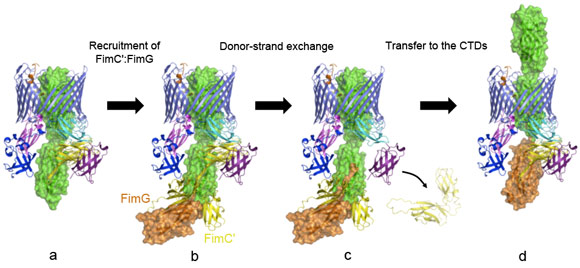
Another striking feature of the active usher conformation is its ability to recruit the next subunit at the NTD site, placing the latter in an ideal position for donor-strand exchange with the subunit bound to the CTDs (the previously incorporated subunit). We conclude that the usher has two binding sites (NTD and CTDs) that act in concert, such that the growing pilus can remain anchored to one, while the next subunit in assembly docks to the other (Figure 2).
These new molecular insights into the pilus biogenesis mechanism offer a new way of stopping bacterial pili growth and so a new approach to the current antibacterial therapy, a promising alternative to face up to the public health crisis caused by increased bacterial resistance to existing antibiotics. Some compounds inhibiting pili formation are already under investigation, but the detailed molecular understanding of the process will help target pilus biogenesis in a more rational manner.
Principal publication and authors
Crystal structure of the FimD usher bound to its cognate FimC–FimH substrate, G. Phan (a), H. Remaut (a,b), T. Wang (c), W.J. Allen (a), K.F. Pirker (a), A. Lebedev (d), N.S. Henderson (e), S. Geibel (a), E. Volkan (f), J. Yan (a), M.B.A. Kunze (a), J.S. Pinkner (f), B. Ford (f,g), C.W. McKay (a,h,i), H. Li (c,j), S. Hultgren (f), D.G. Thanassi (e) and G. Waksman (a,i), Nature 474, 49-53 (2011).
(a) Institute of Structural and Molecular Biology, University College London and Birkbeck College (UK)
(b) Structural & Molecular Microbiology and Structural Biology Brussels, Vrije Universiteit Brussels (Belgium)
(c) Biology Department, Brookhaven National Laboratory, Upton (USA)
(d) Department of Chemistry, University of York (UK)
(e) Center for Infectious Diseases and Department of Molecular Genetics and Microbiology, Stony Brook University (USA)
(f) Department of Molecular Microbiology and Center for Women’s Infectious Disease Research, Washington University School of Medicine, Saint Louis (USA).
(g) Department of Pathology and Immunology, Washington University, St Louis (USA)
(h) London Centre for Nanotechnology, University College London (UK)
(i) Research Department of Structural and Molecular Biology, University College London (UK)
(j) Department of Biochemistry and Cell Biology, Stony Brook University (USA)
References
[1] G. Waksman and S.J. Hultgren, Nature Review Microbiology 7, 765-767 (2009).
[2] H. Remaut et al., Cell 16, 640-652 (2008).
[3] M. Nishiyama et al., EMBO Journal 24, 2075-86 (2005).
Article written by Gilles Phan and Gabriel Waksman
Top image: Secretion of a protein by the chaperone/usher pathway, first step provided by crystal structure.