- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2000
- Life Sciences
- Determination of the Structure of the Capsid of Human Hepatitis B Virus
Determination of the Structure of the Capsid of Human Hepatitis B Virus
Hepatitis B virus is a human pathogen of major significance, as chronic infection can lead to cirrhosis and primary liver cancer, resulting in over a million deaths each year. The viral capsid is composed of a single polypeptide chain, MW 21000, known as the core antigen. When the gene for the core antigen is expressed in Escherichia coli, the expressed protein assembles into shells, which are indistinguishable from capsids isolated from intact virus. The capsid protein contains a very basic C-terminal sequence that is believed to interact with the viral nucleic acid. Removal of this C-terminal sequence by truncation at residue 149 leads to higher levels of bacterial expression with no obvious change in the morphology of the particle. The truncated capsids with T = 4 icosahedral symmetry (Figure 1) have been crystallised and diffraction data were collected to 3.3 Å resolution from 20 cryocooled crystals on beamline ID2. Exposure times of up to 120 seconds for a 0.3° oscillation were required.
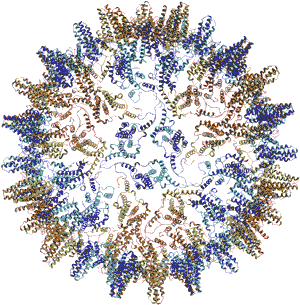 |
Fig. 1: Ribbon representation of the T = 4 hepatitis B capsid, viewed down the icosahedral 3-fold axis.
|
Initial phases were obtained from a 7.4 Å resolution map determined by single particle electron cryomicroscopy. As the capsid packs in the C2 unit cell with a viral 2-fold axis coincident with the crystallographic 2-fold, only one orientational parameter was unknown, and this could be found from the self-rotation function. Initial phases to 8 Å resolution calculated from the EM model were extended to 3.3 Å resolution by exploiting the 30-fold non-crystallographic symmetry. The path of the polypeptide backbone in the resulting electron density map was very clear for residues 1-142 in all four independent copies that make up the icosahedral repeat unit of the capsid. An atomic model was refined with XPLOR imposing strict icosahedral symmetry, giving a final R-factor of 27.1%.
As expected from both CD spectra and the EM work, the monomer fold is largely helical. This is unusual for viral coat proteins, which more typically form a jellyroll or ß-barrel structure. The fold of the monomer is stabilised by an extensive hydrophobic core. Two monomers associate to give a compact dimer in which the two a-helical hairpins pack to form a four-helix bundle. As recognised in the electron cryomicroscopy model, it is this four-helix bundle that forms the characteristic spikes on the surface of the capsid. The C-terminal regions (aa 125-142) of the truncated protein associate around the icosahedral 5-fold and 2-fold (quasi 6-fold) axes and provide most of the packing interactions between adjacent subunits that build up the capsid. The resulting capsid consists of a thin spherical protein shell, with an inner radius of 130 Å and a thickness of 20 Å, from which the four-helix bundles protrude, forming spikes approximately 25 Å in length and 20 Å in width. The capsid is fenestrated, with large pores (approximately 15 Å in diameter) around the icosahedral 2-fold (quasi 6-fold), 3-fold and quasi 3-fold axes. As reverse-transcription of the viral pregenomic RNA to partially double stranded DNA occurs within the assembled capsid, it has been proposed that the pores are essential to allow nucleotides access to the capsid interior during DNA synthesis.
Principal Publication and Authors
S.A. Wynne, R.A. Crowther, and A.G.W. Leslie, Molecular Cell, 3, 771-780 (1999).
MRC, Cambridge (UK)



