- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2000
- Life Sciences
- Structure of a Low-affinity Penicillin-binding Protein from Enterococcus Faecium
Structure of a Low-affinity Penicillin-binding Protein from Enterococcus Faecium
Penicillin-binding proteins (PBPs) are membrane proteins involved in the final stages of peptidoglycan synthesis and represent the main target for b-lactam antibiotics. Enterococcus faecium strains are resistant to penicillin through the overproduction of low-affinity penicillin-binding protein PBP5 [1]. Only one structure of a penicillin sensitive PBP has been solved (PBP2x from Streptococcus pneumoniae [2]). The availability of the structure of a resistant PBP such as PBP5 is of the utmost importance for the understanding of the resistance mechanism developed by the bacteria. It should lead to the development of new antibiotics designed to specifically bind to this protein active site.
Multiwavelength anomalous dispersion seemed the most appropriate technique for deriving phase information in the determination of the PBP5 structure. Small crystals of the SeMet protein were obtained. Data collection was performed under cryogenic conditions at beamline ID14-4. MAD data were collected at three different wavelengths (peak, inflection, remote). At each wavelength, 540 frames were collected with an exposure time of 0.5 s and an oscillation of one third of a degree. The three data sets were complete up to 2.4 Å, a resolution that could only be reached on ID14-4. The space group was C2 (a = 79.3 Å; b = 128.8 Å; c = 236.1 Å and ß = 93.92°) with three molecules in the asymmetric unit (Figure 4). PBP5 is a 647 amino-acid protein with 13 methionines. 30 out of 39 possible selenium sites have been localised using Shake-and-Bake. This was further refined by the use of the MLPHARE program from the CCP4 suite. The MAD map calculated after density modification was suitable to initiate the protein construction. Refinement was completed using CNS.
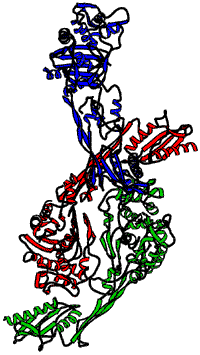 |
Fig. 4: Ribbon representation of the PBP5 from Enterococcus faecium showing the three molecules in the asymmetric unit.
|
PBP5 is composed of two domains. The N-terminal domain folding is new and no biological function has yet been attributed to this domain. The C-terminal domain resembles the transpeptidase domain of the penicillin-sensitive PBP2x and more generally to penicillin-recognising enzymes (DD-peptidases, DD-carboxypeptidases, ß-lactamases of class A, C or D). The active site is delimited by the classical motifs conserved for these proteins.
References
[1] W. Zorzi, X.Y. Zhou, O. Dardenne, J. Lamotte, D. Raze, J. Pierre, L. Gutmann and J. Coyette, J. Bacteriol., 178, 4948-4957 (1996).
[2] S. Pares, N. Mouz, Y. Petillot, R. Hackenbeck and O. Dideberg, Nat. Struct. Biol., 3, 284-289 (1996).
Authors
E. Sauvage (a), G. Leonard (b), P. Charlier (a), R. Herman (a), P. Stefanic (a), J. Coyette (a), Y. Taburet (c), J. Dumas (c), D. Prévost (c), B. Schoot (c) and J.P. Marquette (c).
(a) University of Liège (Belgium)
(b) ESRF
(c) Hoechst Marion Roussel / Aventis, Paris (France)



