- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2008
- Soft condensed matter
- The molecular basis of the braking action of muscle studied by X-ray interference
The molecular basis of the braking action of muscle studied by X-ray interference
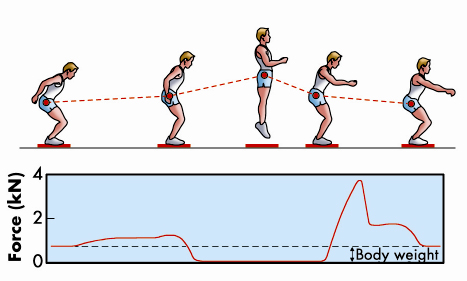
We normally think of muscles as the motors that drive the movements of the body, but they can also act as brakes to resist an external force. An everyday example of this is the action of the muscles in the front of the legs to resist the force of gravity while walking down stairs. A more dramatic example would be deceleration of the body on landing after a jump (Figure 61), when the brakes must be applied in a small fraction of a second. This rapid braking response to an external force is an intrinsic property of isolated skeletal muscle cells. Our recent results at beamline ID02 have revealed its molecular basis for the first time.
 |
|
Fig. 61: The leg extensors decelerate the body after a jump (adapted from Cerretelli, Fisiologia dell’Esercizio, SEU, 2001). |
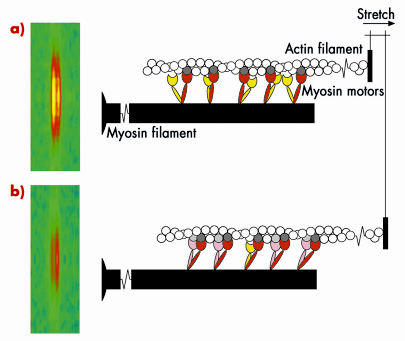
Skeletal muscle cells are packed with parallel arrays of two kinds of filaments, composed of the proteins myosin and actin respectively. Myosin is the motor protein; biochemically driven changes in the shape of part of the myosin molecule, called its motor domain, drive muscle contraction. The actin filaments form the molecular tracks for these myosin motors; when a muscle contracts the myosin motors pull the actin filaments along the myosin filaments. The filaments have an almost crystalline structure; motor domains are spaced at regular intervals along the myosin filaments, and this regular structure produces a measurable diffraction pattern when an isolated muscle fibre is illuminated by a narrow beam of X-rays. One of the X-ray reflections, called the M3, has two closely spaced peaks, Figure 62 (left); this fine structure is an interference effect related to the bipolar structure of the myosin filament: each myosin filament actually contains two arrays of myosin molecules. This X-ray interference effect has proved to be a powerful technique to reveal molecular mechanisms in muscle contraction because it can measure the movements of the myosin motors along the filaments with angstrom resolution in an intact single muscle cell.
 |
|
Fig. 62: Molecular basis of muscle braking; the M3 X-ray reflection and the conformation of the myosin motors before a), and after being stretched b). |
Measurements of these X-ray interference signals with sub-millisecond time resolution were previously used to investigate the molecular basis of force generation and active shortening in the myosin motor [1-3]. We have now used the technique to determine the mechanism of the braking response of muscle to a rapid stretching movement. The results showed that the response of a muscle to being stretched is fundamentally different to that during force generation or active shortening. An important clue to the mechanism came from high resolution mechanical measurements, which showed that the stiffness of the muscle fibre increases in less than a millisecond after being stretched quickly. The integrated mechanical/structural mechanism was revealed by combining the mechanical and X-ray interference results, and taking into account the fact that each myosin molecule has two motor domains. Before being stretched (Figure 62a), one of the motor domains (red) of each myosin molecule is attached to an actin monomer (dark grey), but its partner motor (yellow) is detached. When the muscle is stretched (Figure 62b), some of the partner motors (pink) attach to the neighbouring actin monomer (light grey) and resist being stretched, with the first motor acting as the strain sensor. This model can explain quantitatively the increase in stiffness and the changes in both interference peaks of the M3 X-ray reflection. It also provides an answer to the longstanding question: ‘why does each myosin molecule in muscle have two motor domains?’ When muscle is working as a motor to generate force and active movement, each myosin uses only one of its two motor domains. The partner domain is held in reserve, to act almost instantaneously as a brake when its attachment is promoted by strain in the first motor domain. In this way muscle resists an external stretching movement while minimising the stress on an individual motor.
Principal publication and authors
E. Brunello (a), M. Reconditi (a), R. Elangovan (a), M. Linari (a), Y.-B. Sun (b), T. Narayanan (c), P. Panine (c), G. Piazzesi (a), M. Irving (b) and V. Lombardi (a), Proc. Natl. Acad. Sci. USA 104, 20114-119 (2007).
(a) Università degli Studi di Firenze (Italy)
(b) King’s College London (UK)
(c) ESRF
References
[1] G. Piazzesi et al. Nature 415, 659-662 (2002)
[2] M. Reconditi et al. Nature 428, 578-581 (2004)
[3] G. Piazzesi et al. Cell 131, 784-795 (2007)



