- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2010
- Structural biology
- How the quorum-quenching N-acyl homoserine lactone acylase PvdQ disrupts bacterial virulence
How the quorum-quenching N-acyl homoserine lactone acylase PvdQ disrupts bacterial virulence
In many Gram-negative bacterial pathogens, virulent behaviour is activated by quorum sensing, a process in which bacteria become aware of each other by the action of diffusible N-acyl homoserine lactone (AHL) messaging compounds. To stop or disrupt quorum sensing, bacteria produce enzymes that break down these AHLs, such that they can no longer be used as signalling molecules. This process, called quorum quenching, can be achieved by the hydrolysis of either the ester bond in the homoserine lactone core of the AHL (performed by lactonases) or of the peptide bond that connects the acyl-chain and the homoserine lactone core (performed by acylases). In view of the increase in bacterial drug resistance, quorum quenching has gained interest as a potential tool in the development of novel antimicrobial strategies.
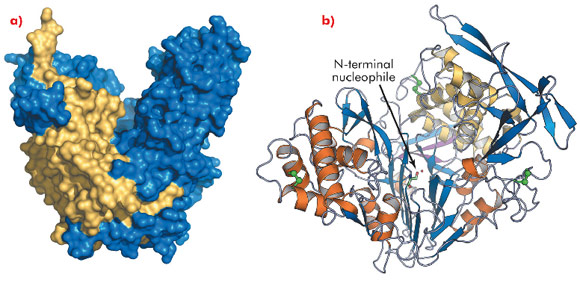
While AHL lactonases have been structurally characterised [1], no data is available for acylases. To gain insights into the mechanism and substrate specificity of AHL acylases, we solved the crystal structure of the AHL acylase PvdQ from Pseudomonas aeruginosa at 1.8 Å resolution, using data collected at ID14-2. PvdQ appears to be an N-terminal nucleophile (Ntn) hydrolase [2] with a heart-shaped structure and a deep crevice in the centre (Figure 121a). The enzyme consists of two peptide chains, with the N-terminal serine of the β-chain, which is located at the bottom of the crevice, forming the catalytic centre (Figure 121b). Near the catalytic centre an extended hydrophobic pocket is buried inside the enzyme core (Figure 122a).
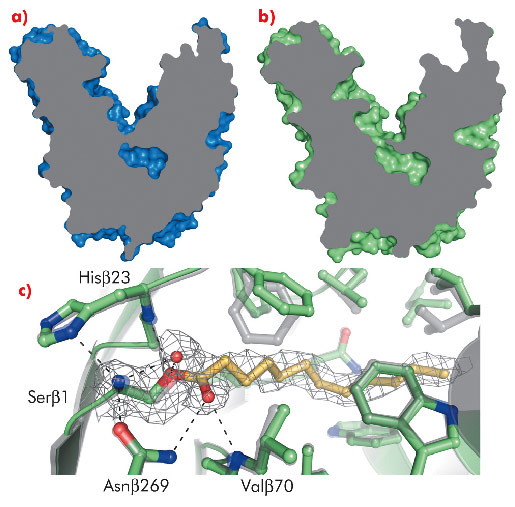
Detailed information on the mechanism behind substrate recognition and catalysis was obtained from PvdQ crystals soaked briefly with C12-homoserine lactone at pH 5.5. This procedure resulted in the formation of an acyl-enzyme intermediate, as revealed by a 2.1 Å resolution crystal structure obtained with data collected at ID14-2(Figure 122c). To accommodate the substrate, the hydrophobic substrate-binding pocket has opened up to solvent (Figure 122b) and the residues lining the pocket have adapted to specifically recognise the long apolar C12 acyl chain of AHL (Figure 122c). This binding mode is in contrast to the AHL lactonases, which specifically recognise the polar homoserine lactone moiety of the AHL. The binding mode of the covalently bound dodecanoic acid indicates a catalytic mechanism of AHL degradation, in which Serβ1 is the nucleophile, Hisβ23 stabilises the reactive state of the nucleophile and Valβ70 and Asnβ269 form the oxyanion hole (Figure 122c).
 |
|
Fig. 122: a) The substrate binding pocket of PvdQ is closed off from the solvent in the unliganded structure; b) it is open when ligand is bound. c) Substrate binding induces subtle conformational changes; the unliganded state is shown in grey, while the structure of the covalently bound dodecanoic acid (yellow) is indicated in green. The important catalytic residues are labelled. |
A BLAST search revealed other Ntn-hydrolases that are homologous to PvdQ, some of which also prefer substrates with long acyl-chain substitutions. PvdQ is the first structurally characterised Ntn-hydrolase that can accommodate such large acyl-chains, and therefore its crystal structure may serve as a paradigm for other Ntn-hydrolases with a similar substrate preference. Moreover, its structure may be used as a template for site-directed mutagenesis to alter the substrate specificity towards short-chain AHLs. This could make the enzyme capable of disrupting quorum sensing-regulated virulence in pathogens that utilise C6-C8 substituted homoserine lactones, for which PvdQ shows only weak activity.
The role of PvdQ in quorum quenching is well established. However, PvdQ is also important for pyoverdine biosynthesis, since pvdQ-gene deletion abrogates production of pyoverdine [3], a siderophore involved in iron homeostasis, which is important for P. aeruginosa survival and virulence. Although the precise role of PvdQ in this latter pathway is not known, Visca et al. have proposed the need for an enzyme that removes a long acyl chain from a pyoverdine precursor [4]. The crystal structures of PvdQ suggest that the distinct binding pocket for long acyl chains might be able to accommodate the acyl chain of a pyoverdine precursor. Whether this specialised acyl-chain binding pocket was initially acquired for AHL hydrolysis or pyoverdine maturation requires further investigation.
Principal publication and authors
M. Bokhove (a), P. Nadal Jimenez (b), W.J. Quax (b) and B.W. Dijkstra (a), PNAS 107, 686–691 (2010).
(a) Laboratory of Biophysical Chemistry, University of Groningen (The Netherlands)
(b) Department of Pharmaceutical Biology, University of Groningen (The Netherlands)
References
[1] D. Liu et al., Proc. Natl. Acad. Sci. USA 102, 11882-11887 (2005).
[2] J.A. Brannigan et al., Nature 378, 416-419 (1995).
[3] P. Nadal Jimenez et al., Microbiology 156, 49-59 (2010).
[4] P. Visca, F. Imperi and I.L. Lamont, Trends Microbiol. 15, 22-30 (2007).