- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2011
- Structural biology
- Nanobody aided crystallisation of β2m amyloidogenic intermediate
Nanobody aided crystallisation of β2m amyloidogenic intermediate
A number of severe contemporary diseases including Alzheimer’s disease, Parkinson’s disease, type 2 diabetes and dialysis related amyloidosis (DRA) are associated with the structural conversion of initially innocuous proteins into insoluble amyloid fibrils. Understanding the molecular basis of this process is crucial for the development of drugs that prevent and/or cure amyloid diseases. However atomic-level investigation of the key intermediates of amyloidogenesis remains one of the supreme challenges of structural biology. This is due to the nature of the process, which may be described as a dynamically evolving equilibrium between large numbers of structural species. These intermediates have highly diverging sizes and are present in uneven amounts and timeframes. β2-microglobulin (β2m) is a well-documented amyloidogenic protein involved in onset of DRA. This type of amyloidosis develops in patients with kidney failure undergoing long-term hemodialysis. Amyloid fibrils are composed mostly of full-length β2m but an N-terminally truncated variant also accumulates in the protein deposits in the musculoskeletal system of DRA patients.
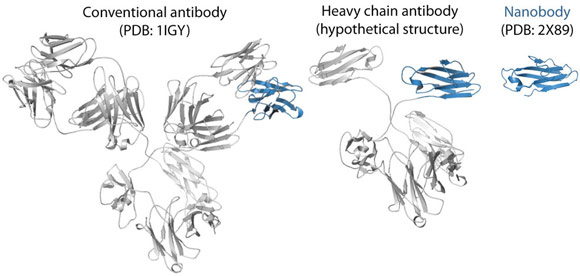 |
|
Fig. 115: Ribbon representations of a conventional antibody, a heavy chain antibody and a nanobody. The variable domain of the heavy chain is depicted in blue. |
To trap and characterise the early intermediates of β2m fibril assembly, we used single domain antibodies. The antigen-binding site of these antibodies consists of a single domain, referred to as VHH or nanobody (Figure 115). Nanobodies are derived from heavy chain only antibodies that along with conventional antibodies naturally occur in camelids [1]. We generated a panel of anti-β2m nanobodies and found that some of the nanobodies stabilise the amyloidogenic β2m variant (∆N6β2m) abolishing its propensity to fibrillise. One of these nanobodies proved to be instrumental in solving the crystal structure of a dimeric intermediate of β2m fibrillation process (Figure 116a). This high-resolution structure (2.2 Å; beamline ID29) confirms the predicted domain swapped nature of the early amyloidogenic intermediate and explains the many local structural perturbations (mutations, deletions, ligand binding) that have been associated with the transition of the soluble β2m into insoluble fibrils. The properties of the swapped dimer provide new insights on the elongation mechanism by self-templated growth, leading to protofibrils and ultimately amyloids. During the self-association of ∆N6β2m, two hinges that correspond to the heptapeptide NHVTLSQ, refold into extended β-strands and stack into a novel two-stranded anti-parallel β-sheet. This heptapeptide forms amyloids in isolation demonstrating that this peptide by itself has a high propensity to form amyloid structure upon exposure [2]. In the newly formed two-stranded sheet, the backbone donor and acceptor sites of Val85 and Leu87 are exposed to solvent (Figure 116b) and are prone to stack with other b-strands in a parallel or antiparallel configuration. Indeed, other strands may associate perpendicular to build large intermolecular β-sheets that run parallel to the axis of the growing oligomers (Figure 116c). It thus appears that the swapped dimer can serve as a structural nucleus for the growth of the cross-β spine of elongating fibrils by templating the hydrogen-bonding network connecting the strands. The remaining core domains may decorate the spine and protect it from solvent.
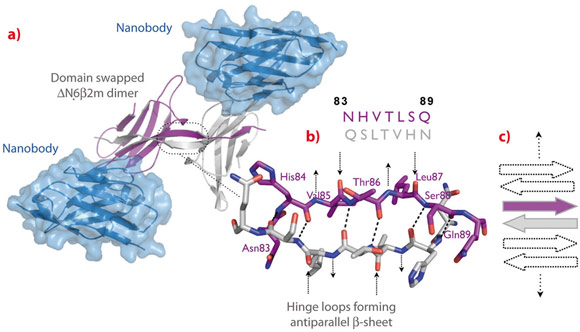 |
|
Fig. 116: Structure of the ∆N6β2m swapped dimer. a) ∆N6β2m swapped dimer stabilised by two nanobody molecules. b) Atomic structure of the open interface of the ∆N6β2m dimer. The main chain NH and CO groups of His84, Thr86, and Ser88 are hydrogen-bonded to the carbonyls and the amides of Ser88, Thr86, and His84 on the adjacent strand, respectively. The backbone donor and acceptor sites of Val85 and Leu87 are exposed to solvent (indicated by arrows). c) Cartoon presenting the possible association of β-strands causing self-templated growth of the fibril core. |
Most remarkably, ∆N6β2m swapped dimer meets many characteristics that have been attributed to prefibrillar intermediates of β2m fibrillogenesis. Phe30, His31 and Pro32, three residues particularly involved in amyloidogenesis are located on the tip of the first loop that connects strands B and C. In the native monomer, parts of all three connecting loops are shielded from solvent by the N-terminal peptide that is missing in ∆N6β2m variant, explaining why the truncated species is less stable and unlike wild-type protein has a higher tendency to self-associate and forms amyloid fibrils even at physiological pH [3].
The identification and characterisation of oligomers preceding the formation of fibrils is of particular interest because evidence is growing that these species are likely to play a critical role in the pathogenesis of protein deposition diseases. The use of nanobodies to trap a single oligomeric intermediate of a dynamically evolving equilibrium between large numbers of conformational species along the amyloidosis pathway may be broadly applicable in the study of other dynamic systems characterised by structural diversity or in the prevention or treatment of amyloid diseases.
Principal publication and authors
K. Domanska (a,b), S. Vanderhaegen (a,b), V. Srinivasan (a,b), E. Pardon (a,b), F. Dupeux (c), J.A. Marquez (c), S. Giorgetti (d), M. Stoppini (d), L. Wyns (a,b), V. Bellotti (d) and J. Steyaert (a,b), Proc Natl Acad Sci U S A, 108, 1314-9 (2011).
(a) Department of Molecular and Cellular Interactions, Vlaams Instituut voor Biotechnologie, Brussels (Belgium)
(b) Structural Biology Brussels, Vrije Universiteit Brussel (Belgium)
(c) Grenoble Outstation, European Molecular Biology Laboratory and Unit of Virus Host-Cell Interactions, Grenoble (France)
(d) Department of Biochemistry, University of Pavia (Italy)
References
[1] Hamers-Casterman et al., Nature 363, 446–448 (1993).
[2] Ivanova et al., Proc Natl Acad Sci USA 103, 4079–4082 (2006).
[3] Esposito et al., Protein Sci 9, 831–845 (2000).



