- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2011
- Structural biology
- Tracking the protein motions that harvest energy
Tracking the protein motions that harvest energy
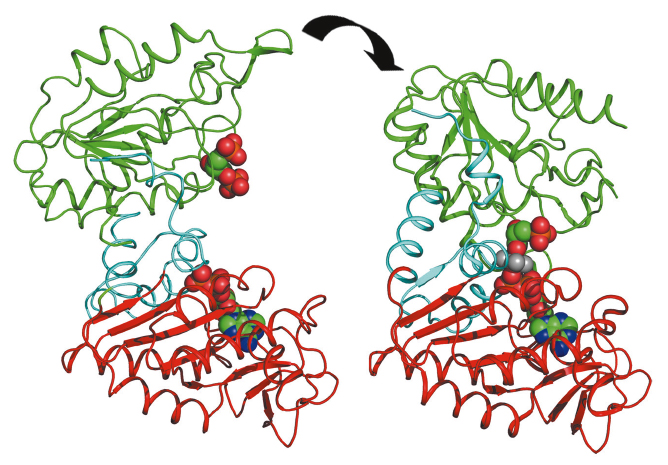
Glycolysis is a metabolic pathway common to all organisms that breaks down sugar to release energy. The pathway is very ancient, does not require oxygen and is likely to have evolved before the accumulation of oxygen in the earth’s atmosphere around 3 billion years ago. Phosphoglycerate kinase (PGK) catalyses the seventh step in glycolysis and is the first enzyme in the pathway to produce energy, rather than consume it, by the creation of adenosine triphosphate (ATP), a molecule that is a universal ‘energy currency’ used to do work in cells. PGK is a target for drugs against anaerobic pathogens, such as the causative agents of sleeping sickness and malaria, that depend solely on glycolysis for energy metabolism. Additionally, the phosphoryl-transfer activity of PGK is important in the processing of anti-retroviral pro-drugs effective against HIV and hepatitis. More recently, it has been found that PGK activity is also used as a signalling mechanism in oncogenesis. A complete understanding of the mechanism of this enzyme is thus essential to improve the processing of pro-drugs and to define its signalling activity in cancer. PGK is composed of two domains that bind two substrates separately (Figure 122). Crystal structures have defined both open and closed conformations for PGK [1] and suggest that a significant closure of the enzyme via a “hinge-bending” mechanism is required for catalysis to occur. By combining high resolution crystal structures solved from data collected on ID14-2 with solution small angle X-ray scattering (SAXS) data collected on ID14-3, the extent and timing of the domain motions that lead to catalysis in PGK have now been elucidated.
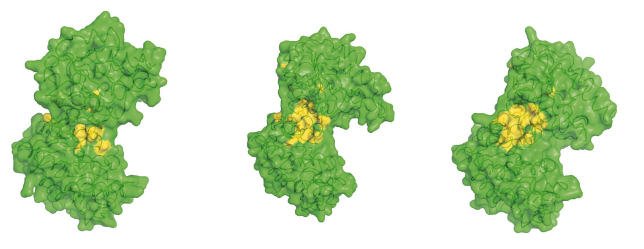
SAXS measurements were used to examine the resting, open state of the enzyme when no substrates are bound as well as various states of the enzyme in catalytic turnover. By comparing the SAXS envelope determined for the protein in solution to the crystal structures we have obtained it was found that the apo-enzyme adopts a more open conformation than previously thought. During catalysis, the relative contribution of each state defined by a crystal structure to a SAXS scattering curve can be obtained. Surprisingly, it was found that, during catalytic turnover, the enzyme spends only 7% of its time in the fully closed, catalytically active conformation and is in the fully open conformation most of the time. In order to define the origin of the stability of the open state, a model of the molecular interactions in this state was required. However, as SAXS data provide structural features to around only 15 Å resolution, the details necessary in such a model are missing. By refining the atomic positions of a crystal structure against SAXS data and applying deformable elastic network (DEN) protocols [2], a molecular model of the fully open conformation observed in solution was obtained. Analysis of this model revealed that a large, solvent-exposed hydrophobic patch present in the closed conformation is masked from solvent in the open conformation (Figure 123). The occlusion of this hydrophobic region from solvent could account for the preference of PGK to adopt an open conformation, as this will be thermodynamically more stable and implies that, while the enzyme constantly moves between the open and closed states, it has an energetic preference for the open conformation. The hydrophobic patch thus acts as a ‘spring’, constantly applying pressure to the closed conformation and leading to efficient release of products after catalysis. A model of cycling and catalysis may therefore be proposed where unliganded PGK remains open (ready to bind substrates, or release products) most of the time. The interpolation of crystal structures and SAXS data has allowed a high resolution time resolved model of catalysis to be proposed.
Principal publication and authors
L. Zerrad (a), A. Merli (b), G.F. Schröder (c), A. Varga (d), E. Graczer (d), P. Pernot (a), A. Round (e), M. Vas (d) and M.W. Bowler (a), J. Biol. Chem. 286, 14040-14048 (2011).
(a) ESRF
(b) University of Parma (Italy)
(c) Forschungszentrum Jülich (Germany)
(d) Hungarian Academy of Sciences (Hungary)
(e) EMBL, Grenoble (France)
References
[1] M.J. Cliff et al. J. Am. Chem. Soc. 132, 6507-6516 (2010).
[2] G.F. Schröder et al. Nature 464, 1218-1222 (2010)