- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2011
- Soft condensed matter
- Snapshots of a photosynthetic reaction centre
Snapshots of a photosynthetic reaction centre
Nature harvests the energy content of light, the major source of energy for the biosphere, primarily via photosynthetic reaction centres. These integral membrane protein complexes are found in plants and photosynthetic bacteria and function by driving the transport of electrons across an energy transducing biological membrane in response to the absorption of a photon by a special pair of chlorophyll molecules.
Using the method of time-resolved Laue diffraction, we have successfully observed structural changes occurring in connection with the primary light driven reactions of a bacterial photosynthetic reaction centre (Figure 64). This work builds upon earlier developments at ID09B where the light driven reactions of myoglobin with bound carbon-monoxide, and photoactive yellow protein, were studied by time-resolved Laue diffraction. The new discovery described here is the first reproducible electron density changes in an integral membrane protein complex.
 |
|
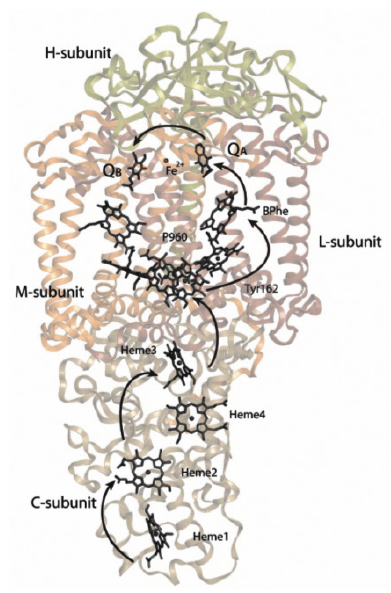
Fig. 64: Overview of the structure and energy transfer reactions of the bacterial photosynthetic reaction centre of Blastochloris viridis. Light causes electrons to move from the special pair (P960) to quinone molecules (QA and QB) on the opposite side of the membrane. P+960 is, in turn, reduced from the tetra-heme cytochrome subunit. |
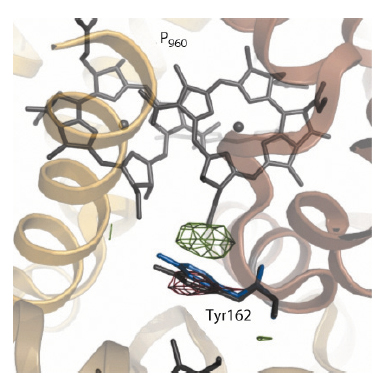
Figure 65 shows the difference Fourier electron density map (red: negative density; green: positive density) captured 3 milliseconds following laser photo-activation of crystals of the reaction centre. Paired positive and negative changes in electron density can be observed that are associated with a fully conserved tyrosine residue (TyrL162). This key residue plays an important functional role by bridging the special pair (P960, which is photooxidised in the primary light conversion event of photosynthesis) to Heme 3 (which reduces P+960).
 |
|
Fig. 65: Difference Fourier electron density changes (red: negative density; blue: positive density) captured by using time-resolved Laue diffraction. TyrL162 is observed to move 1.3 Å (grey: resting conformation; blue: photo-activated conformation) towards the special pair (P960) 3 milliseconds following photo-activation. |
These experimental observations were interpreted as due to a change in the conformation of TyrL162, which moves approximately 1.3 Å towards the special pair (Figure 65: blue: conformation photo-activated state; grey: conformation resting state) in response to light. Molecular dynamics simulations and free energy calculations suggested that this motion arises because Tyr162 becomes deprotonated following the primary electron transfer event of an electron away from the special pair (P960). This supports the concept of complementary electron-proton transfer reactions, whereby the excess positive charge on P+960 induces a proton transfer away from TyrL162 in a direction opposite to that of the electron transfer. This principle may find application in the future design of energy transducing systems of artificial photosynthesis.
Principal publication and authors
A.B. Wöhri (a,b), G. Katona (c), L.C. Johansson (c), E. Fritz (c), E. Malmerberg (c), M. Andersson (a), J. Vincent (d), M. Eklund (d), M. Cammarata (e,f), M. Wulff (e), J. Davidsson (d), G. Groenhof (g) and R. Neutze (c), Science 328, 630-633 (2010).
(a) Department of Chemical and Biological Engineering, Chalmers University of Technology, Göteborg (Sweden)
(b) Present address: AstraZeneca Research and Development, Cell, Protein, and Structural Sciences, Mölndal (Sweden)
(c) Department of Chemistry, Biochemistry and Biophysics, University of Gothenburg, Göteborg (Sweden)
(d) Department of Photochemistry and Molecular Science, Uppsala University (Sweden)
(e) ESRF
(f) Present address: SLAC National Accelerator Laboratory, Menlo Park, California (USA)
(g) Computational Biomolecular Chemistry Group, Department of Theoretical and Computational Biophysics, Max Planck Institute for Biophysical Chemistry, Göttingen (Germany)



