- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2012
- Structural biology
- Structural dynamics of the aminoacylation and proof-reading functional cycle of bacterial leucyl-tRNA synthetase
Structural dynamics of the aminoacylation and proof-reading functional cycle of bacterial leucyl-tRNA synthetase
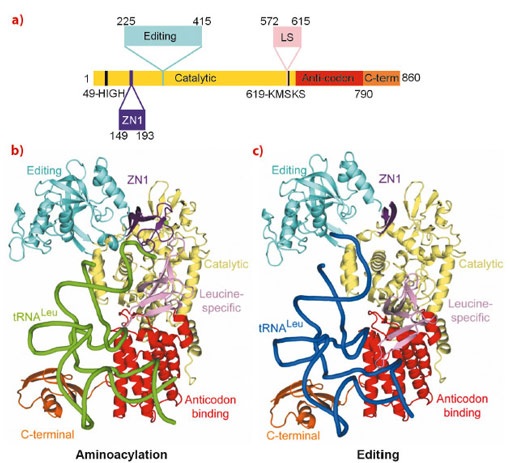
The aminoacyl-tRNA synthetases are responsible for attaching a particular amino acid to its cognate transfer-RNA. The accuracy of this reaction is essential to the fidelity of protein synthesis and thus for cell survival. However, distinguishing structurally related amino acids with sufficient accuracy presents a fundamental challenge for the molecular recognition of the aminoacylation active site. To overcome this problem, at least six different aminoacyl-tRNA synthetases, for example LeuRS, IleRS and ValRS, have developed highly evolved, proof-reading and editing mechanisms, which reduce the error rate to better than 1 in 3000. For example, LeuRS possesses a robust editing mechanism to hydrolytically deacylate tRNALeu that has been mischarged with non-cognate amino acids such as isoleucine or methionine. LeuRS, a ~100 kDa protein, comprises a synthetic enzyme body including the Rossmann-fold catalytic domain, which activates the amino acid with ATP and then ligates it to the tRNA 3’ end, and the anticodon binding domain. In addition, there are four flexibly linked domains, denoted zinc (ZN1), editing, leucine-specific (LS) and C-terminal (Figure 23a). Proof-reading requires that the tRNA 3’ end, initially charged (or mischarged) in the synthetic site, translocates to the editing site, located ~35 Å away in the independently folded editing domain. Although the LeuRS complex with the tRNA bound in the editing site (‘editing state’) has been previously reported, structural information about the aminoacylation state and how the tRNA translocates between the two sites, and about the roles of each of the externally linked domains, was lacking.
Making extensive use of the ERSF high intensity beamlines, we have now determined the crystal structure of the functional aminoacylation complex of E. coli LeuRS (LeuRSEC) at 2.5 Å resolution. In this ternary complex, the 3’ end of E. coli tRNALeu is bound in the synthetic site and poised to interact with leucyl-adenylate (present as a non-hydrolysable analogue), the intermediate aminoacylation reaction (Figure 23b). To complement this, we have also determined the crystal structure of the same system at 2.1 Å resolution but with the tRNA 3’ end bound in the editing site (Figure 23c). Comparison of the two structures reveals substantial domain and active site rearrangements that accompany tRNA translocation between the two functional conformations (Figure 24; also see the movie: http://www.cgl.ucsf.edu/chimera/animations/tRNA/leucyl-tRNA.mov). A hypothesis for the mechanism of translocation is as follows. We suppose that the aminoacylation state is a high energy state, under strain due to the unusual conformation of the 3’ end of the tRNA, the distorted state of the adenylate and the fully closed conformation of the catalytic ‘KMSKS’ loop (Figure 24a). Formation of this state is possible due to a high binding energy of the tRNA 3’ end as a result of its numerous contacts to various domains of the enzyme, including the editing domain. Once the covalent integrity of the adenylate is broken by transfer of the amino acid moiety to the tRNA, the strain is released by relaxation of the KMSKS loop into its open conformation, dissociation of AMP and the flip of the leucine-specific domain (Figure 24b). Concomitantly, the KMSKS loop, the ZN1 and editing domain lose their interactions with the tRNA, destabilising the binding of the acceptor end of the now charged tRNA. This allows the tRNA 3’ end to flip to its preferred low energy conformation in the editing active site (Figure 24b). Globally, the editing domain slightly rotates as the tRNA pivots around its fixed contact points on the anti-codon binding domain and the C-terminal domain moves synchronously, maintaining its contacts to the tRNA. In this conformation the tRNA has far fewer contacts and is already partially disassociated from the catalytic domain. This favours total tRNA release in the case of correctly charged Leu-tRNALeu or possibly direct re-aminoacylation, without full release, of hydrolysed and recycled, mischarged tRNA.
 |
|
Fig. 24: Schematic diagram of the structural changes between the aminoacylation (a) and proof-reading (b) conformations of the LeuRSEC-tRNALeu complex. Colour code as in Figure 23 (dotted regions are disordered in the crystal structure). Leucine is represented as a white oval and AMP as a black rectangle. The black arrows in (a) indicate direction of movement during translocation to the editing state. |
Our results provide new insight into the dynamic mechanism of aminoacylation and proof-reading in LeuRS, a molecular machine essential for accurate protein synthesis.
Principal publication and authors
A. Palencia (a), T. Crépin (a), M.T. Vu (b), T.L. Lincecum (b), S.A. Martinis (b) and S. Cusack (a), Nat Struct Mol Biol. 19, 677-84 (2012).
(a) European Molecular Biology Laboratory (EMBL), Grenoble Outstation and Unit of Virus Host-Cell Interactions, University of Grenoble-EMBL-Centre National de la Recherche Scientifique, Grenoble (France)
(b) Department of Biochemistry, University of Illinois,Roger Adams Laboratory, Urbana (USA)