- Home
- Users & Science
- Find a beamline
- Structural biology
- Our beamlines
- BM29 BioSAXS
- Beamline setup
- Sample requirements
Sample requirements
BioSAXS sample requirements
Here are the important physicochemical factors to control for a convenient sample preparation for BioSAXS experiments.
Concentration series
To obtain X-ray scattering data which can be used to draw valid biological conclusions, it is important to follow a rigorous experimental protocol of sample preparation. Following are some recommendations for a proper sample preparation.
Monodisperse samples (no aggregation)
Prior your visite, it is indispensable to well-purify the samples (gel-filtration, chromatography, dialise, etc) and check their monodispersity using Dynamic Light Scattering (DLS) or Multi-Angle Laser Light Scattering (MALLS) techniques.
Analytical Ultracentrifugation technique (AUC) can also be used to determine the sizes, the molecular masses and the oligomeric state of biological samples .
The data quality will depend strongly on the level of sample purity, a purity level close to those of crystallography, i.e. greater than 90%.
Concentration serie (at least 3) for each sample
To determine if there are any inter-particle effects experimentally (aggregation / repulsivion), we typically measure a series of concentrations for each macromolecule of interest (e.g. 1, 2, 5 and 10 mg/ml).
To ensure correct normalization of the scattering intensity for the differences in the protein concentration measurement of the sample concentration is required (accuracy better then 10% is needed.
Prior to SAXS data collection, we recommand to centrifuge the samples for few minutes at 4 deg. C and recuperate only the supernatant. After that one should re-measure the absorbance (optic density, OD) at 280 nm for determining sample concentrations (knowing extintion its coefficient). A Nanodrop Spectrophotometer and a Desktop Centrifuge are available in the Sample Preparation lab next to the beamline for this purpose.
Buffer requirements
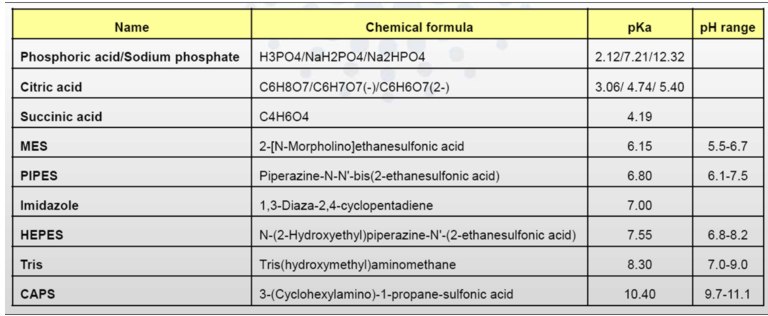
To obtain the scattering data from macromolecule of interest, the scattering from the buffer in which a given protein is dissolved must be measured (better if both before and after each sample measurement). Most of biological buffers are compatible with SAXS studies, see Table. Excess volumes of buffer should be prepared for these measurements (in mL range).
Table 1. Most frequently used buffers (salts) of biological samples
The buffer composition should exactly match the protein solvent. Any DTT, glycerol, detergent (DDM or SDS), small molecules (ATP/ADP) or salts added into samples, must be also put into buffers!
Pay also attention to the quantity of salts or surfactants added into samples. It is recommended to use:
- Ionic strength of salts (NaCl, HCl, Na2SO4, K2HPO4 ...) < 1M
- Concentration of small proteins (ADP or ATP) < 10 mM
- Glycerol percentage < 30% (usually, in the range [5-10%])
- Detergent concentration < CMC (Critical Micelle Concentration)
Particular Case of Detergents
Detergents are used with membrane lipids and membrane proteins to increase their solubility and stability in solution. However, we recommend not using detergents in the entire protein preparation for SAXS measurements because they affect the protein signal by micelles’ formation.
One should i) go for SEC online with SAXS measurement or ii) produce buffers which match free micelles concetration around samples for each sample concetration (can be quite tricky).
Required Sample Volumes
Manual mode:
in some cases, the sample needs to be loaded manually to protect the fragile 3D structure of the protein (e.g, the fibrilled proteins or silk-proteins). For such studies, we recommend 50 µL of sample per measurement . Note that this is the volume for an individual measurement and depending on the analyzed data, additional measurements at other concentration may be required for a comprehensive data set to be collected.
The sample is loaded either using a syringe or using the sample changer application (by dragging up from the well to the capillary). In both cases, the speed of the sample loading requires to be low (1-2 µL/s).
~ 3 concentrations in hour are measured in manual mode.
Sample changer modes
a) Static mode measurement (fixed liquid position)
The illuminated volume of the sample is about 1 µL, however in order to avoid the scattering from interfaces sample/air which completely overhelmes the measured data is recommended to load at least 4 µL into the capillary. Note, that this is the volume inside the exposure capillary, however to allow the needle to suck it from the sample container (a PCR tube for example) more than this is needed depending on sample viscosity, let say 10-15 µL should be in the tube.
b) Flow mode measurement (regular and homogeneous motion of the sample inside the capillary)
This mode can prevent/help with radiation damage, recommended is to load about 30 µL (50 µL even better) and to use "flow" mode which allows the sample moves through the beam during data collection to hit always fresh sample. Note that this is our defaut mode.
About 20 concentrations in hour are measured with the robot.
Generally, if you have 100 µL of stock solution you are safe. Use 50 µL for first measurement and dilute other 50 µL to the middle concentration (and have so 100 µL of it), again use 50 µL of it for measurement and remaining 50 µL dilute again (and again)...
SEC online requirements
- at least 10 µL of sample (50 or 100 µLbetter) with highest concentration you can (2-5 mg/mL are good);
- bring you own column(s);
- enough buffer (in L range) which match your sample, good if filtered and outgassed (can be re-done in sample preapration lab prior to the experiment);
- some knowledge of a strandard SEC/HPLC system and software.