- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2014
- Structural biology
- Crystal structure of a common GPCR-binding interface for G-protein and arrestin
Crystal structure of a common GPCR-binding interface for G-protein and arrestin
The large family of G-protein-coupled receptors (GPCRs) controls various sensory and physiological responses and is thus a major focus of research and of interest for medical drug targets. These receptors bind a variety of ligands (e.g. light-depending retinal, hormones, opioids, odorants, peptides, neurotransmitters) and communicate such extracellular signals into the cell by coupling to intracellular heterotrimeric G-proteins (Gαβγ) and arrestins (Figure 113) [1]. In G-protein-signalling, the Gα C-terminus (GαCT) binds to a cytoplasmic crevice formed in the agonist-bound active receptor [2,3], eventually catalysing GDP/GTP exchange in the Gα-subunit (1). Another binding partner is arrestin, whose canonical role is to stop signalling by blocking receptor coupling to G-proteins, or to initiate arrestin-dependent signalling by bringing together other signalling proteins. In our studies, we have addressed the question of how arrestins employ the cytoplasmic crevice within the 7 transmembrane (TM) bundle of the active receptor for the high-affinity interaction necessary to block G-protein binding or stimulate arrestin mediated signalling [4,5]. This work was motivated by our observation of a consensus sequence motif, (E/D)x(I/L)xxxGL (Figure 113, inset left), which is conserved in GtαCT of the Gi/Gt-family and the “finger loop” (ArrFL) from the central crest region of all four subgroups of arrestin [6].
 |
|
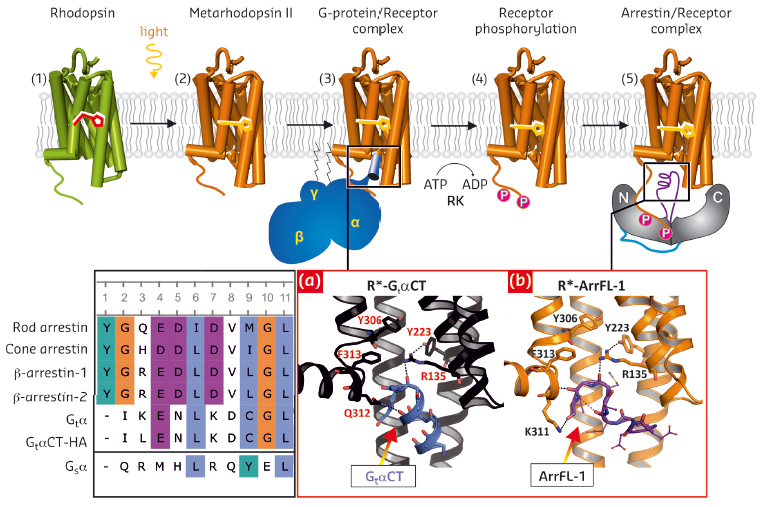
Fig. 113: Overview of rhodopsin signal transduction and deactivation. (1) Dark-state bovine rhodopsin with inverse agonist 11-cis-retinal. (2) Light-activation and formation of metarhodopsin II with agonist all-trans-retinal. (3) Coupling to the G-protein. (4) Phosphorylation of the activated receptor by its specific kinase. (5) Arrestin binding to the phosphorylated receptor. Inset (left): Sequence alignment of a proposed common sequence motif: rod photoreceptor arrestin (Arr1; residues 68–77), cone photoreceptor arrestin (Arr4; 63–72), β-arrestin-1 (Arr2; 64–73), β-arrestin-2 (Arr3; 65–74), wildtype GtαCT (340–349), high-affinity variant GtαCT-HA (340–349), Gsα (385–394). Inset (right): View of the cytoplasmic receptor crevice from the crystal structures depicted as ribbon-stick diagrams of a), R*- GtαCT (coloured in black-blue, PDB entry 3DQB, [2]) and b) R*- ArrFL-1 (coloured in orange-purple, PDB entry 4PXF, [6]). |
We have used protein X-ray crystallography, Fourier-transform-infrared spectroscopy, functional competition assays and UV-Vis absorption spectroscopy on solutions (“Extra-Meta II assay”) and on crystals (at beamline ID29S – The Cryobench) to gain insight into how an active receptor binds to ArrFL. Firstly, we co-crystallised the active receptor rhodopsin R* (as light-activated metarhodopsin II or as the active form of the apo-protein opsin, Ops* [1,2,3,6]) with three different ArrFL peptides derived from rod photoreceptor arrestin, β-arrestin-1 and β-arrestin-2. The crystal structure of active rhodopsin with a peptide analogue derived from ArrFL-1 (rod photoreceptor arrestin), was solved at 2.75 Å resolution using diffraction data collected at beamline ID29. ArrFL binds to the cytoplasmic crevice of R* with a similar reverse turn structure as observed for the C-terminal GtαCT [2]. In both cases, a hydrogen bond is formed between the ArrFL-1 reverse turn and Arg135 from the highly conserved E(D)RY motif of R*.
To verify the functional relevance of the observed peptide binding to the receptor crevice, we applied three different functional assays to native disc membranes filled with rhodopsin. These functional experiments showed similar R* binding properties of Gi/Gt and ArrFL peptides. The key player, Arg135, essential for receptor activation and G-protein-coupling, has also a decisive role in arrestin binding (Figure 113, right). However, significant differences between ArrFL and GtαCT are seen at the rim of the receptor crevice. In ArrFL-1, the interaction with TM5/6 is partially replaced by interactions with TM7/H8 (specifically to the conserved NPxxY(x)5,6F motif) and directed towards the intracellular loop 2 (CL2).
Overall, the crystal structure indicates a general mechanism of arrestin binding and the remarkable homology seen between GtαCT and ArrFL suggests that the (E/D)x(I/L)xxxGL motif evolved in both G-protein and arrestin to allow recognition and interaction with the same binding crevice presented by the active GPCR [6]. Despite these similarities, we also identified pronounced differences, namely a larger hydrophobic interaction surface in the case of GtαCT, which may contribute to the distinct biological functions of G-proteins and arrestin.
Principal publication and authors
M. Szczepek (a,b), F. Beyrière (a), K.P. Hofmann (a), M. Elgeti (a), R. Kazmin (a), A. Rose (a), F.J. Bartl (a), D. von Stetten (c), M. Heck (a), M.E. Sommer (a), P.W. Hildebrand (a) and P. Scheerer (a,b), Nature Communications 5, 4801 (2014).
(a) Institute of Medical Physics and Biophysics, Charité - Universitätsmedizin Berlin (Germany)
(b) Institute of Medical Physics and Biophysics, Group Protein X-ray Crystallography and Signal Transduction, Charité - Universitätsmedizin Berlin (Germany)
(c) ESRF
References
[1] K.P. Hofmann et al., Trends Biochem Sci. 34, 540-552 (2009).
[2] P. Scheerer et al., Nature 455, 497-502 (2008).
[3] H.-W. Choe et al., Nature 471, 651-655 (2011).
[4] A.K. Shukla et al., Nature 497, 137-141 (2013).
[5] Y.J. Kim et al., Nature 497, 142-146 (2013).
[6] M. Szczepek et al., Nature Communications 5, 4801 (2014).



