- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2017
- Structural biology
- A new model for the activation of the immune system
A new model for the activation of the immune system
A new model has been suggested for the activation of an important branch of the innate immune system. The activation of the large C1 protein complex is a fundamental mechanism in immunology, and this new model is likely to spur developments in immunotherapy with the aim of fighting cancer and infections.
An important part of the immune system is the so-called complement system. When the immune system detects a microorganism or other molecular signs of danger, the C1 complex is converted into an active enzyme that can cleave other proteins, thus initiating a cascade of proteolytic events. The end result of this reaction is that invading pathogenic microorganisms are more efficiently ingested by immune cells. In addition, an inflammatory response is triggered, leading to the elimination of the microorganisms. In the past few years, there has been increased focus on the C1 complex, since – in addition to its function in the immune system – it has been shown to be closely involved in the development of the nervous system and neurological disorders. One of the most exciting recent findings is that the C1 complex participates in trimming of the synapses mediating signalling between neurons.
The C1 complex circulates in an inactive state in blood and other extracellular fluids. Immunology research has so far suggested that activation of the C1 complex is due to a complicated structural change within each individual C1 complex. Such alteration of the structure was proposed to occur when the C1 complex binds to the danger-presenting object, e.g., a pathogenic organism or a dying human cell. The underlying assumption was that the C1 complex has a compact conformation that allowed the protease subunits within each individual complex to activate each other through an intramolecular activation mechanism, but this compact conformation has not yet been observed.
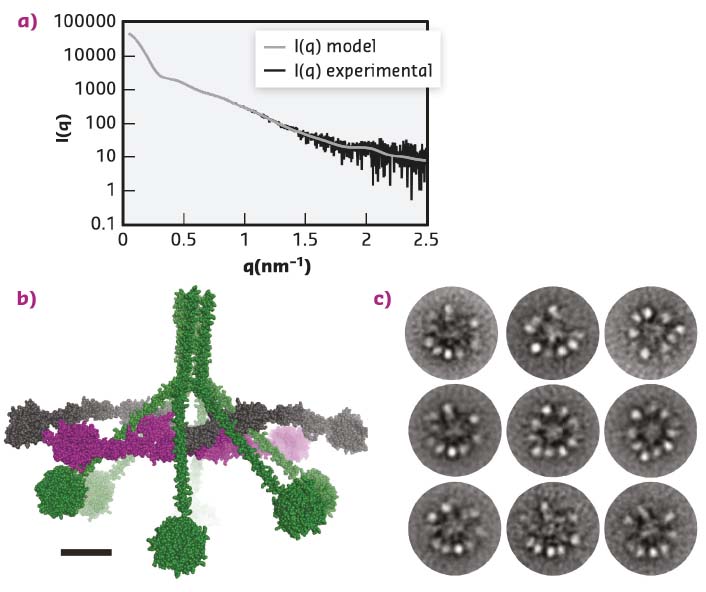
The MBL:MASP-1 complex resembles the C1 complex but is smaller and functions in a different branch of the complement system. Functional and structural studies showed that activation of the MBL:MASP-1 complex is an intermolecular reaction where neighbouring complexes activate each other and that the MBL:MASP-1 complex has an extended conformation with the catalytically active protease domains located at the periphery of the molecule. Whether the C1 complex also has such an extended conformation or indeed adopts a compact conformation was unknown. As the C1 complex is highly non-globular and flexible, crystallisation was deemed unlikely, instead the C1 complex and its sub-component C1r2s2 were investigated in parallel with small-angle X-ray scattering (SAXS) and Electron Microscopy (EM). SAXS data were collected for the C1 complex at BM29 and at PETRA-III, while the data for the C1r2s2 were collected at BM29. For the SAXS studies, measuring the C1 complex eluting directly from a size exclusion column made it possible to remove the signal from contaminating or aggregated molecules, and was crucial for improving the quality of the data obtained. After data collection, ab initio modelling was used to produce a molecular envelope, showing that the C1 complex was hollow and not a compact particle. The ab initio model was further supported by rigid body modelling that resulted in atomic models of the C1 complex that fitted the SAXS data (Figure 17a-b). All these models had the protease domains present in the periphery of the C1 complex and not tightly packed in the centre of the molecule. EM micrographs confirmed this finding (Figure 17c).
 |
|
Fig. 17: The structure of the C1 complex. a) SAXS data recorded at PETRA-III, similar data were obtained at BM29. The grey curve displays how a curve calculated from the model (b) fits the experimental data. c) The ‘class averages’ of the C1 complex images recorded with electron microscopy. The overall architecture suggested by microscopy confirms the model obtained from the SAXS data presented in (b). |
Since these results, obtained by two orthogonal methods, suggest that the proteolytically active domains are located in the periphery and not in the core, the existing model of C1 activation as an intramolecular process needs to be reconsidered. Instead, activation is likely when two C1 complexes are located sufficiently close to each other that their protease subunits may activate each other in an intermolecular reaction. This model is also in agreement with the results obtained for the homologous MBL:MASP-1 complex [1,2].
The biomedical implication of the new C1 complex activation model is that it may be sufficient to tether the C1 complex to a cancer or pathogen cell in a defined overall orientation in order to elicit an innate immune response towards a malicious cell. If this is true, a specific recognition event of molecular patterns on the surface of the offending cell is not required, and binding of the C1 complex to cells may turn out to be an attractive new approach to immunotherapy.
Principal publication and authors
Structure and activation of C1, the complex initiating the classical pathway of the complement cascade, S.A. Mortensen (a), B. Sander (b), R.K. Jensen (c), J. Skov Pedersen (d), M.M. Golas (a), J.C. Jensenius (a), A.G. Hansen (a), S. Thiel (a) and G.R. Andersen (b), PNAS 114, 986-991 (2017); doi: 10.1073/pnas.1616998114.
(a) Department of Biomedicine, Aarhus University (Denmark)
(b) Department of Molecular Biology and Genetics, Aarhus University (Denmark)
References
[1] T.R. Kjaer et al., Structure (London, England: 1993) 23, 342 (2014).
[2] S.E. Degn et al., PNAS 111, 13445-13450 (2014).



