- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2017
- Structural biology
- Revealing the mechanism of bacterial transmembrane signalling
Revealing the mechanism of bacterial transmembrane signalling
Crystal structures of an E. coli sensor protein in ligand-bound and ligand-free states reveal the mechanism of transmembrane signalling employed by bacterial sensor histidine kinases.
Cellular perception of external stimuli is an ability of every living organism that is critical for survival. Sensor histidine kinases (HKs) are a large class of membrane receptor proteins that, in bacteria, are involved in different processes including pathogenicity and cell growth. This makes them attractive targets for study, particularly as various HKs of different species are expected to share a similar mechanism due to a conserved domain architecture and dimeric working state. However, despite the fact that HKs are widespread and despite the availability of some structural information, there has been no consensus concerning the molecular mechanism of sensory signal transduction through the membrane. The reason is that the available structural information is either of low resolution or from isolated domains of HK molecules, thus giving little insight into the concerted structural transformations associated with signal transduction.
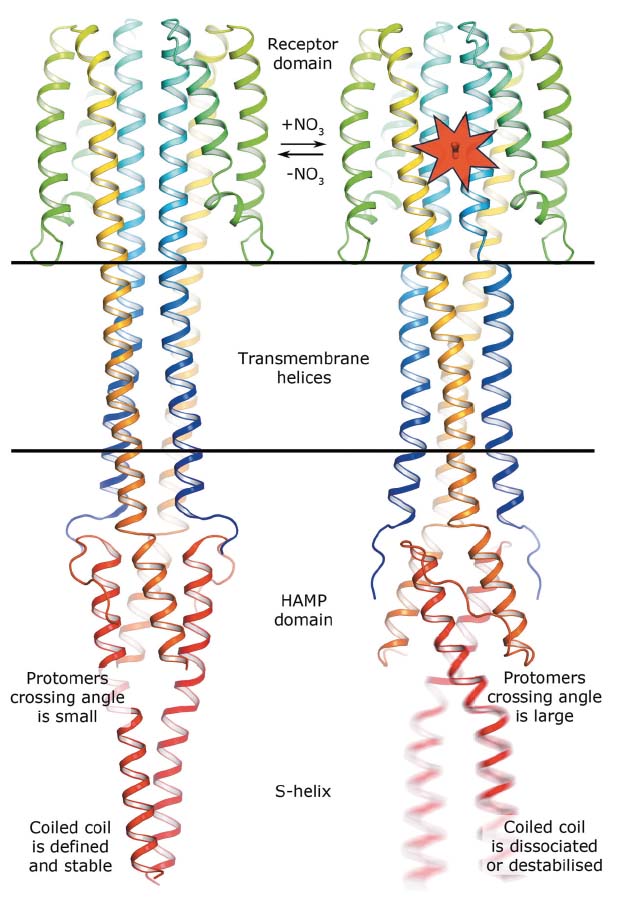
In meso crystallisation and single-wavelength anomalous dispersion experiments at beamline ID23-1 were used to determine the crystal structures in both ligand-bound and ligand-free states of a truncated construct of the Escherichia coli nitrate/nitrite sensor HK, NarQ. In E. coli, NarQ senses nitrate and nitrite ions in the periplasm and regulates the metabolism of these. This is very important as bacteria use these ions as electron acceptors in the respiratory chain in the absence of oxygen. Comparing the structures obtained reveals that binding of nitrate or nitrite to the periplasmic sensor module of NarQ causes a structural rearrangement, resulting in a symmetric displacement of ~2.5 Å of two of the four transmembrane α-helices in the HK dimer. On the cytoplasmic side, this relatively small displacement is transmitted to the amplifying HAMP domain, which undergoes a lever-like motion that results in the maximum displacement amplitude of 7 Å at the end of the HAMP output helices, likely destabilising the coiled coil phase (see Figure 10). The latter phenomenon is assumed to impact the stability of downstream helices, thus modulating the efficiency of kinase function.
 |
|
Fig. 10: Mechanism of NarQ transmembrane signalling. Binding of the ligand launches a series of structural changes including a piston-like relative displacement of the transmembrane helices towards the periplasm, consequent lever-like conformational changes in the HAMP domain and the dissociation or destabilisation of the signalling helix. |
The structural similarities between HKs strongly suggest that the piston-like displacement in the transmembrane domain is an important facet in signal transduction by all HK proteins. The mechanism of the signal amplification seen for the NarQ HAMP domain also creates a framework for understanding the principles of signalling of all HAMP-containing proteins. Overall, the results show how a mechanistic signal is generated and amplified while being transduced through HKs over distances of 100 Å or more. The study may provide insights for the further understanding of signalling pathways in different species of microorganisms and, potentially, promote the production of new antimicrobial treatments other than conventional antibiotics.
Principal publication and authors
Mechanism of transmembrane signaling by sensor histidine kinases, I. Gushchin (a,b), I. Melnikov (c), V. Polovinkin (a,b,d), A. Ishchenko (a,e), A. Yuzhakova (a,b), P. Buslaev (b), G. Bourenkov (f), S. Grudinin (g,h,i), E. Round (a,d), T. Balandin (a), V. Borshchevskiy (a,b), D. Willbold (a,j), G. Leonard (c), G. Büldt (b), A. Popov (c) and V. Gordeliy (a,b,d), Science 356, 6342, eaah6345 (2017); doi: 10.1126/science.aah6345.
(a) Institute of Complex Systems (ICS), ICS-6: Structural Biochemistry, Research Centre Jülich (Germany)
(b) Moscow Institute of Physics and Technology, Dolgoprudniy (Russia)
(c) ESRF
(d) Université Grenoble Alpes, CEA, CNRS, IBS, Grenoble (France)
(e) Institute of Crystallography, University of Aachen (RWTH) (Germany)
(f) European Molecular Biology Laboratory (EMBL), Hamburg Outstation (Germany)
(g) Université Grenoble Alpes, LJK, Grenoble (France)
(h) CNRS, LJK, Grenoble (France)
(i) Inria, Grenoble (France)
(j) Institute of Physical Biology, Heinrich Heine University, Düsseldorf (Germany)



