- Home
- News
- Spotlight on Science
- Bio-SAXS at ID14-3:...
Bio-SAXS at ID14-3: looking at proteins in their native state – a valuable complement to protein crystallography studies
24-06-2011
The complete automation of data collection is maximising the potential of bio-SAXS at the ESRF. Never before have SAXS studies of protein in solution been so easy. Academics and industrialists are getting excellent results and some are even using the beamline by remote operation.
Share
Small-angle X-ray scattering (SAXS) has experienced a renaissance in recent years for structural biology, in part because it allows the study of biological macromolecules, from individual proteins to multi-subunit complexes, in solution under nearly physiological conditions. However, SAXS beamlines at European synchrotrons have been highly oversubscribed with proposals from a variety of different scientific fields. The need to increase access and efficiency led to a collaboration between the EMBL and the ESRF to convert ID14-3 from a crystallography beamline into a dedicated bio-SAXS beamline in 2008.
The trilateral collaboration involving the EMBL's Grenoble and Hamburg outstations together with the ESRF has lead to significant improvements in throughput, ease of use and data quality through the development and deployment of an automated environment for samples in solution. As is the case with macromolecular crystallography (MX), automation has increased throughput and reliability. It has also enabled remote operation (the first remote experiment was performed on the 4th March 2011) and facilitated service access for industrial customers (the first service data collection was performed on the 18th April 2011). The liquid handling features of the sample changer unit (SCU) allows pipetting for dilution or additions of material prior to loading micro lit re volumes of samples (with concentration measurement using an in-line spectrometer). This automated sample environment is presented in Figure 1.
 |
|
Figure 1. The new automated sample environment in place on ID14-3. |
All aspects of the experiment, including sample loading, data collection and cleaning can be controlled by the beamline control GUI (BsxCuBE) for fully-automated data acquisition. Automated cleaning is much more thorough than can be achieved manually, greatly increasing stability and reliability of background measurements and therefore data quality. When combined with feedback from the automated data analysis tools developed by EMBL Hamburg, users can be more rigorous and obtain reliable data with a greater degree of confidence under a range of physiologically relevant conditions (pH 3 - 12, temperature (4 – 60ºC), salts, ligands, etc.).
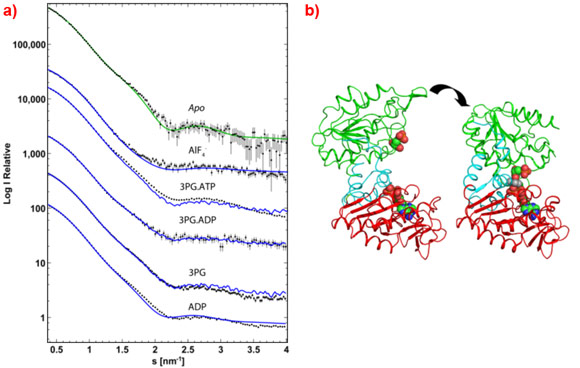
The ease of collecting data from a large range of conditions allows the functional states of active systems to be probed in order to elucidate the physiological processes. Recently, this has enabled the definition of a high resolution, time resolved model of catalysis by phosphoglycerate kinase [1]. SAXS data (Figure 2a) collected at ID14-3 were used to verify various fixed points within the reaction cycle (Figure 2b) and analyse the enzyme in catalytic turnover to show that the complex is mostly in an open conformation . This allows the substrates access to the complex which closes only for catalysis when it brings the substrates together, after which the complex springs open to release the reaction products and is then ready to repeat the cycle.
The ability to combine different structural techniques (MX, NMR and EM) with solution scattering data enables functional analysis under a wide range of experimental conditions offers exciting opportunities to the user community. With further collaboration between ILL and ESRF via the PSB SAS platform, users can maximise the benefits of visiting the EPN campus, collecting complementary data using neutrons and X-rays during the same trip.
The bio-SAXS beamline at ID14-3 is now more than a place to collect data, it is a much needed and valuable tool which not only encourages collaborative research but when used in combination with results from other techniques, can greatly increase their relevance and impact.
Principal publication and authors
P. Pernot (a), P. Theveneau (a), T. Giraud (a), R. Nogueira Fernandes (b), D. Nurizzo (a), D. Spruce (a), J. Surr (a), S. McSweeney (a), A. Round (b), F. Felisaz (b), L. Foedinger (b), A. Gobbo (b), J. Huet (b), C. Villard (b) and F. Cipriani, J Phys: Conf Ser 247, 012009-1-012009-8 (2010).
(a) ESRF
(b) EMBL, Grenoble (France)
References
[1] L. Zerrad, et al., J Biol Chem. 286, 14040-14048 (2011).
Top image: Robot handling proteins in solution.