- Home
- News
- Spotlight on Science
- The structure of...
The structure of Wza reveals a new class of membrane protein
10-12-2006
Wza, a carbohydrate exporter that is part of a much larger molecular machine, was the first lipoprotein shown to traverse the outer membrane. The crystal structure of Wza, finally solved after 6 years effort and numerous data collection trips to the ESRF, shows the protein to comprise 4 domains and that the transmembrane domain folds as a helical barrel. This is a novel membrane spanning motif for bacteria and the structure solution raises the possibility that other proteins assigned as lipoproteins may, in fact, also be transmembrane proteins.
Share
Outer membrane lipoproteins, which have their leader sequence cleaved off and a thioacyl glycerol attached to the newly created N-terminal cysteine, were known to bind to the inner leaflet but were not thought to traverse the outer membrane. The first such protein for which it was clearly demonstrated that it did so was Wza, a protein responsible for the export of the carbohydrate capsule from gram negative bacteria [1]. Wza sits in the outer membrane and is thought to be part of a large molecular machine (Figure 1).
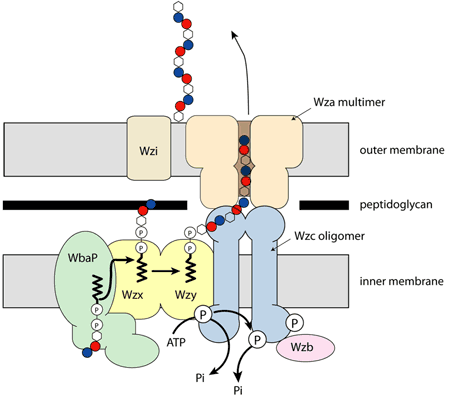 |
|
Fig. 1: Wza in the cellular context. |
A structural study of the protein was initiated some six years ago. Wza turned out to be straightforward to express but as always finding the right crystallisation conditions took considerable trial and error. The successful structure determination that followed relied on regular and frequent access to the ESRF insertion device-based MX beamlines. Of the many trips made over a five year period, only one yielded the structure. Collection of high quality diffraction data proved challenging and crystal decay due to radiation damage was a serious hindrance. Another problem was that most crystals diffracted to around dmin = 3.0 Å. This meant the most important and interesting data for structure solution was collected at the edges / corners of the tiled CCD, a region of such detectors known to be higher in noise. After 14 unsuccessful attempts incremental improvements in crystal quality resulted in a single-crystal diffracting to dmin = 2.26 Å. Data collected from this crystal finally allowed us to locate the selenium atoms in a selenomethionyl derivate of the protein from a peak wavelength SAD experiment and structure solution quickly ensued.
The structure of Wza fully rewards the long years and beam time expended upon it. The protein has four domains, arranged around a central 8 fold rotational axis and its structure is best described as an amphora (Figure 2). Analysis of the amino acid sequence of Wza had failed to identify any transmembrane regions. However, examination of the protein surface reveals that the helical barrel (domain R4, Figure 2) is hydrophobic indicating that it is the membrane spanning region. The size of the hole through the helical barrel is consistent with its role in carbohydrate export. The protein is closed in the periplasm by the PES domain, the defining sequence feature of this protein family. The other two domains are gene duplications and share structural similarity to ubiquitin (P. Artymiuk personal communication). The protein is proposed to bind Wzc (Figure 1) which opens the PES domain and thus allows carbohydrate export.
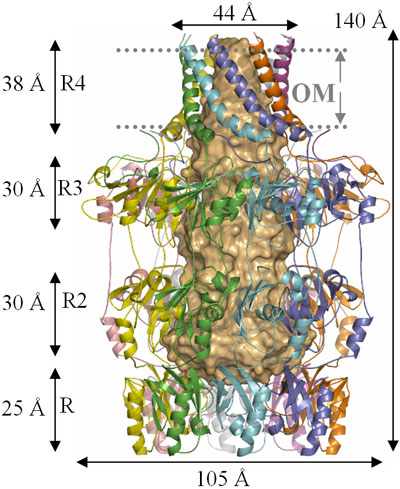 |
|
Fig. 2: The structure of Wza (reproduced from Nature [2]). |
The helical barrel is a novel membrane spanning motif (previously bacterial outer membrane proteins were thought to use only β-barrels for this purpose), and raises the possibility that other lipoproteins may also, in fact, be transmembrane proteins, but mis-annotated because amino acid sequence analysis techniques have not identified the signature.
The project was funded by the Wellcome Trust and the experimental structural biology carried out by the Scottish Structural Proteomics Facility (a BBSRC / SFC funded project). Gordon Leonard, Dominique Bourgeois and David Hall from the ESRF provided considerable support over the years.
References
[1] J. Drummelsmith, C. Whitfield, Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane, EMBO J. 19, 57-66 (2000).
[2] C.J. Dong, K. Beis, B.R. Clarke, A. Brunkan, C. Whitfield, J.H. Naismith, Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein, Nature, 444, 226-230 (2006).
Authors
C. Dong (a), K. Beis (a), J. Nesper (b), A.L. Brunkan-LaMontagne (a), B.R. Clarke (b), C. Whitfield (b) and J.H. Naismith (a).
(a) Centre for Biomolecular Sciences, The University of St Andrews, Fife (UK)
(b) Department of Molecular and Cellular Biology, The University of Guelph, Ontario (Canada)



