- Home
- News
- Spotlight on Science
- Snapshot of a synaptic...
Snapshot of a synaptic complex formed during DNA double-strand break repair
01-04-2008
The mechanisms by which DNA damage occurs and how this damage is repaired is fundamentally important in the field of cancer biology and has wide implications in many other fields.
Share
DNA double-strand breaks (DSBs) are a potentially lethal form of cellular damage as failure to repair such breaks can lead to genome instability, a potent inducer of cancer development. In human cells, the non homologous end-joining (NHEJ) pathway is critical for the repair of DSBs. A functionally homologous, but much simpler, repair system also exists in bacterial cells. A UK-based team led by Dr Aidan Doherty at the University of Sussex in Brighton, in collaboration with groups in Spain and at the ESRF, have targeted the key proteins involved in this repair process in Mycobacterium tuberculosis to gain an understanding of how this fundamental process works. Their findings, published in the journal Science [1] describe novel biochemical and structural studies that provide insights at the molecular level into the mechanism of DNA double-strand break repair in prokaryotes.
In eukaryotic cells, DSB repair processes are orchestrated by the NHEJ complex, a DNA repair apparatus composed of many cellular proteins. In comparison, the picture is much less complex in bacterial cells because the NHEJ complex comprises only two proteins, one of which is a multi-functional repair ligase (DNA ligase D) that can process fragmented ends and synthesise a new DNA linkage. Much work has been carried out on the activities and structure of these bacterial repair ligases, however, a molecular understanding of how bacteria mechanistically orchestrate DSB repair using just two proteins remains to be established. In particular, still unknown is how the broken ends of the DNA are brought together, processed and “stitched” back together.
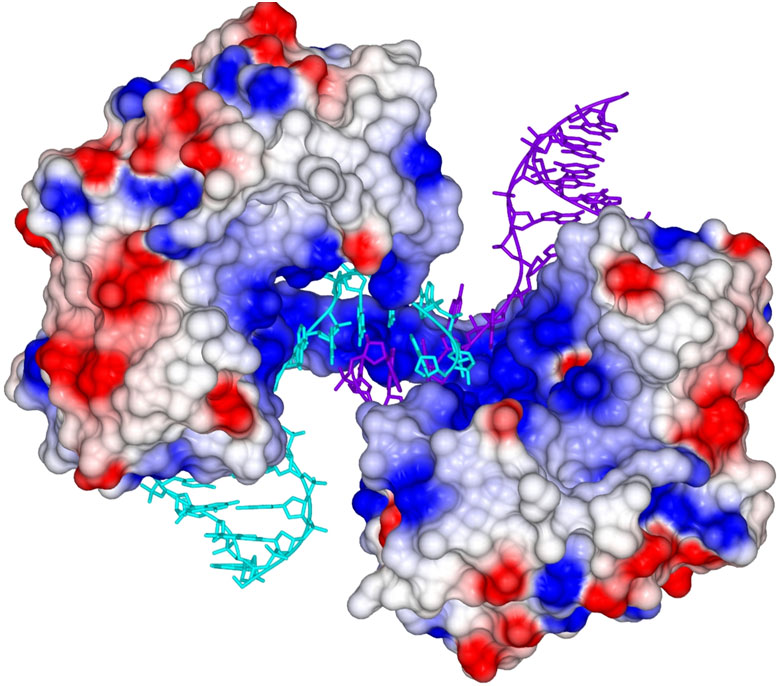
The Sussex-based team combined biochemical, biophysical and structural techniques to unravel the role played by the polymerase domain of LigD (PolDom) in mediating synapsis formation. An initial crystal structure of the PolDom protein alone, was solved by a MAD (multiple-wavelength anomalous diffraction) experiment carried out on the CRG beamline BM16 at the ESRF. This structure was then used as a search model to solve the structure of a large binary crystallographic complex formed between PolDom and DNA containing a double strand break.
The structure of this polymerase-mediated DNA repair complex shows how two molecules of PolDom can directly co-ordinate the association of the ends of DNA breaks, forming a large synaptic complex in which end-processing and ligation can subsequently occur (Figure 1). The structure provides a molecular snapshot of a key intermediate step in the break repair process, suggesting that these enzymes play a pivotal role in end-bridging. These findings will enable scientists studying DNA break repair to draw parallels and consider the possible implications in closely related eukaryotic systems, where the mechanisms that govern the co-ordination of NHEJ processes are poorly understood.
Principle publication and authors
N.C. Brissett (a), R.S. Pitcher (a), R. Juarez (b), A.J. Picher (b), A.J. Green (a), T.R. Dafforn (c), G.C. Fox (d), L. Blanco (b), A.J. Doherty (a), Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science 318, 456 (2007).
(a) Genome Damage and Stability Centre, University of Sussex, Brighton (UK)
(b) Centro de Biología Molecular Severo Ochoa, CSIC-UAM, Madrid (Spain)
(c) Department of Biosciences, University of Birmingham (UK)
(d) BM16, ESRF