- Home
- News
- Spotlight on Science
- How the genetic...
How the genetic code is accurately translated into protein
09-11-2009
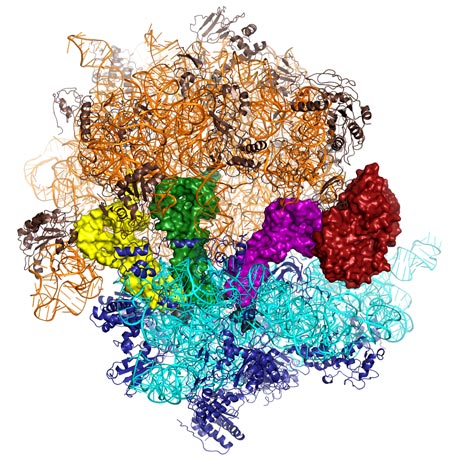
The production of proteins is performed in all organisms by ribosomes, nanomachines that carry out the translation of an mRNA sequence and the subsequent synthesis of a protein from that sequence. Ensuring that the genetic code is correctly translated into a functional protein is one of the most important steps that ribosomes perform in protein synthesis. A new crystal structure of the ribosome, in complex with an elongation factor, reveals how the ribosome guarantees that the genetic code has been correctly recognised before allowing peptide synthesis to occur.
Share
DNA contains the information needed to produce the proteins that perform nearly all the functions in the body from breaking down food to muscle contraction. The first step in the process of producing proteins is to transcribe the DNA sequence into a template (called messenger ribonucleic acid, mRNA) that is sent into the cell. Nanomachines called ribosomes read this template and use the information contained in it to ligate together a precise sequence of amino acids to produce functional proteins. Ribosomes are themselves mostly made of RNA, with some protein, and are comprised of a large and a small subunit (Figure 1). The small subunit closes around a sequence of mRNA and it is at this point that the process of translation and peptide synthesis starts. The ribosome uses aminoacyl-tRNA (transfer RNA with individual amino acids attached) as substrates. Each tRNA matches the three letter genetic code to a specific amino acid thus allowing a sequence of amino acids to be attached to each other in the order dictated by the genetic code. Aminoacyl-tRNA is delivered to the ribosome by a protein called Elongation Factor-Tu (EF-Tu). This protein binds tRNA tightly and will only release it when the genetic code has been accurately translated. This process has been the focus of intense research for many years, and many antibiotics target EF-Tu. While many hypotheses have been proposed, the mechanism of how EF-Tu functions with the ribosome to ensure accuracy in protein synthesis remained unknown without a detailed molecular picture from X-ray crystallography.
Venki Ramakrishnan’s group from the Laboratory of Molecular Biology in Cambridge (UK) managed to crystalise the entire ribosome in complex with EF-Tu and aminoacyl-tRNA. Using a powerful antibiotic (kirromycin), that prevents EF-Tu from releasing tRNA, the complex was stabilised sufficiently to form an ordered crystal lattice. The extreme size (~30 nm in the largest dimension) and complexity of the ribosome/EF-Tu/aminoacyl-tRNA system leads to an enormous heterogeneity in crystal quality. This means that the diffraction properties of hundreds of crystals needed to be examined in a synchrotron X-ray beam in order to select those that diffract to a sufficient resolution to allow proper visualisation of the interactions between the molecules. This type of sample evaluation is only possible at the ESRF Structural Biology beamlines due to their high level of automation of beam alignment and sample handling. The final data sets were collected on beamline ID14-4, where the low divergence of the beam is extremely useful when collecting data from crystals with large unit cell dimensions.
The crystal structures reveal a complicated transmission of information from the centre of the ribosome to the peripherally bound EF-Tu, a distance of 80 Å. When EF-Tu delivers tRNA to the ribosome it is also bound to a molecule called GTP. If the genetic code is correctly translated this causes conformational changes in the tRNA and EF-Tu. This rearrangement leads to the chemical breakdown of GTP, which acts like a switch holding EF-Tu to tRNA. When the switch is tripped, the aminoacyl-tRNA can enter the active site and the amino acid enters the growing polypeptide chain. The antibiotics used in this study trap EF-Tu in the state when the switch is being tripped providing an exquisite molecular snapshot of the process of translation.
Principal publication and authors
T.M. Schmeing, R.M. Voorhees, A.C. Kelly, Y-G. Gao, F.V. Murphy, J.R. Weir, and V. Ramakrishnan, The Crystal Structure of the Ribosome Bound to EF-Tu and Aminoacyl-tRNA, Science 326, 688-694 (2009).
MRC Laboratory of Molecular Biology, Cambridge (UK)
Article written by M.W. Bowler, ESRF.
Top image: How the genetic code is accurately translated into protein (Image credit: Larissa Ulisko, Rebecca Voorhees, Martin Schmeing)