- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2015
- Structural biology
- Exposing the ‘Achilles heel’ of dengue virus serotypes
Exposing the ‘Achilles heel’ of dengue virus serotypes
Candidate Dengue disease vaccines have yet to offer protection against all four serotypes of dengue virus. Recently, human antibodies that neutralise all four serotypes have been isolated. Subsequent structural investigations show that these bind to a serotype-invariant site on the surface of the virus, revealing an ‘Achilles heel’ of dengue virus and potentially paving the way for the development of vaccines giving protection against all dengue virus serotypes.
Dengue disease is spread to humans by mosquito bites and is caused by four serotypes of the dengue virus (DENV). Of the nearly 400 million people per year infected with the virus, 75% exhibit no symptoms and recover quickly. However, for a small percentage, symptoms include bleeding from the nose or gums, severe abdominal pain, persistent vomiting and breathing difficulties. Although Phase III trials of potential vaccines have recently been carried out, current candidate vaccines show only partial protection with none providing protection against serotype 2 [1, 2]. Recently a series of human antibodies (bnAbs) that neutralise all four serotypes has been isolated [3] and structural studies of several of these bnAbs in complex with the envelope glycoprotein E (‘protein E’) from DENV serotype 2 were carried out using diffraction data collected at ESRF beamlines ID23-2 and ID29 and at SOLEIL. Protein E, relatively conserved among different serotypes, contains two conserved N-linked glycosylation sites (N67, N153) and is the only target of antibodies which neutralise dengue virus. In the mature dengue virus, 90 protein E dimers, arranged in an icosahedral configuration, completely coat the surface of the virus. Once the virus enters into cells, an irreversible conformational change leads to the fusion of viral and endosomal membranes.
 |
|
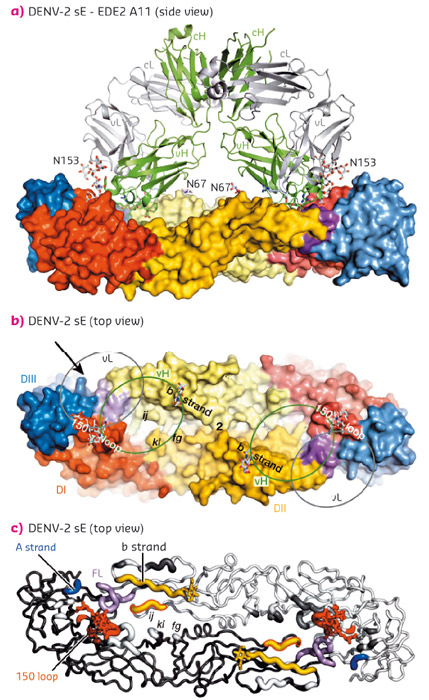
Fig. 136: Interactions between the bnAbs and dengue virus protein E dimers. (a) Complex with bnAbs A11. The protein E dimer (side view) is shown in a surface mode representation coloured according to the domain structure shown in (b). The two N-linked glycans are shown as ball-and stick. A11 is shown in ribbon representation (heavy and light chains in green and grey respectively). (b) The unliganded Protein E dimer (2-fold axis labelled ‘2’). The ovals vH and vL show the contact sites of heavy and light chains of A11 respectively. (c) ‘Worm’ representation of the regions of the protein E dimer targeted by all four bnAbs. The main segments of the epitope are coloured and labelled. Reprinted by permission from Macmillan Publishers Ltd: Nature 520, 109-113 (2015), copyright 2015. |
The crystal structures of four bnAbs/Protein E dimer complexes were obtained, showing that all four bnAbs (B7, C8, C10 and A11) straddle the protein E dimer interface and interact with protein E dimers in very similar regions, conserved in all four serotypes of dengue virus (Figure 136). On one subunit of the dimer, these regions include the N67 glycan bearing b strand, the ij loop and the domain II fusion loop. On the opposing subunit, the 150 loop is either directly targeted or rendered disordered by interaction of the antibody with domains I and III of protein E.
 |
|
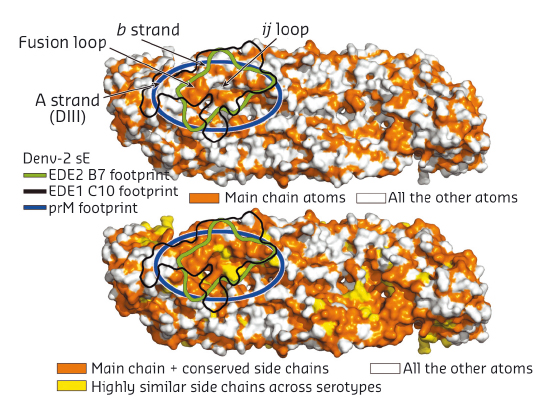
Fig. 137: The bnAbs protein E dimer epitope. The epitopes of bnAbs C10 (black line) B7 (green line) are indicated. The interaction area of the precursor membrane glycoprotein prM is shown as a blue oval. Reprinted by permission from Macmillan Publishers Ltd: Nature 520, 109-113 (2015), copyright 2015. |
The major basis of the action of the bnAbs studied here seems to be interaction, in the specific context of an intact protein E dimer, with the domain II fusion loop and its surrounding area. This latter observation is important. In infected cells, immature DENV virions containing 180 copies of a heterodimer of protein E with the precursor membrane glycoprotein prM bound at their surface bud into the endoplasmic reticulum lumen, where the pH is neutral. Upon transportation to the acidic external medium, prM protects protein E from undergoing a premature acid-induced fusogenic conformational change and remains bound to protein E dimers until the immature virions reach the exterior of the cell where the pH is neutral. Here, prM is released allowing the pre-fusion protein E dimers to become fusogenic upon encountering a new cell. The protein E dimer epitope (EDE) recognised by the bnAbs studied here maps extremely well with the binding site of prM on the protein E dimers (Figure 137), which thus represents an ‘Achilles heel’ of DENV as it is conserved across all four serotypes. A possible strategy for the development of vaccines against DENV might therefore be based on the presentation of DENV pre-fusion E dimers to the immune system with the aim of producing a response to the E dimer epitope identified here.
Principal publication and authors
Recognition determinants of broadly neutralizing human antibodies against dengue viruses. A. Rouvinski (a,b), P. Guardado-Calvo (a,b), G. Barba-Spaeth (a,b,c), S. Duquerroy (a,b,c), M-C. Vaney (a,b), C.M. Kikuti (a,b), M.E. Navarro Sanchez (a,b), W. Dejnirattisai (d), W. Wongwiwat (d), A. Haouz (e), C. Girard-Blanc (e), S. Petres (e), W.E. Shepard (f), P. Desprès (a), F. Arenzana-Seisdedos (a), P. Dussart (g), J. Mongkolsapaya (d,h), G.R. Screaton (d) and F.A. Rey (a,b,e), Nature 520, 109-113 (2015); doi: 10.1038/nature14130.
(a) Institut Pasteur, Paris (France)
(b) CNRS UMR 3569, Paris (France)
(c) Université Paris-Sud, Faculté des Sciences, Orsay (France)
(d) Division of Immunology and Inflammation, Department of Medicine, Imperial College, London (UK)
(e) CNRS UMR 3528, Paris (France)
(f) Synchrotron SOLEIL, Gif-sur-Yvette (France)
(g) Institut Pasteur de Guyane, Cayenne (French Guiana)
(h) Dengue Hemorrhagic Fever Research Unit, Siriraj Hospital, University, Bangkok (Thailand)
References
[1] M.R. Capeding et al., Lancet 384, 1358–1365 (2014).
[2] D. Normile, Science 345, 367–368 (2014).
[3] W. Dejnirattisai et al., Nature Immunol. 16, 170–177 (2015).



