- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2016
- Structural biology
- United LEAFY binds, alone it falls
United LEAFY binds, alone it falls
The LEAFY protein, a transcription factor responsible for flower development, is able to assemble itself into small chains made up of several proteins. This mechanism allows it to bind to and activate regions of the genome that are inaccessible to a single protein.
Flowering plants, angiosperms, represent the vast majority of plant species and are the main basis for agriculture. The flower, which contains both male and female organs in close proximity, is a very efficient structure for reproduction and in part helps to explain the impressive success of this group of plants in evolution. The protein orchestrating the development of flowers is called LEAFY [1]. It encodes a plant specific transcription factor that binds DNA using a unique fold based on a helix-turn-helix motif [2].
LEAFY also contains a second conserved domain whose function had remained elusive. Using an integrated structural biology approach, the function of this region was shown to be an oligomerisation domain and a crystal structure was obtained following data collection at beamline ID23-2, which showed it to belong to the sterile alpha motif (SAM) family [3]. In vitro experiments showed that the domain helps LEAFY to bind DNA containing multiple binding sites. This property was confirmed in vivo in plants expressing either the wild-type protein or an engineered variant that can no longer oligomerise. More unexpectedly, it was also found that the oligomerisation domain is required for LEAFY to bind to closed chromatin regions, which are normally poorly accessible to transcription factor binding. Figure 75 shows a dimer of LEAFY bound to DNA.
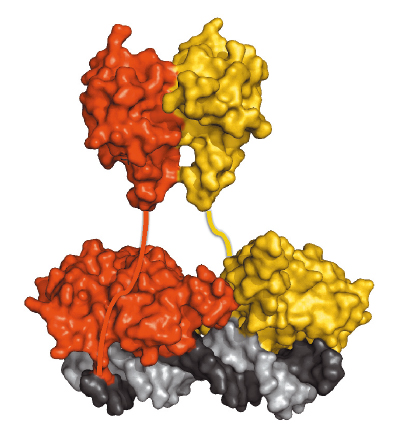 |
|
Fig. 75: A dimer of LEAFY sitting on DNA. The helix-turn-helix domain is in direct contact with DNA whereas the SAM oligomerisation domain is represented above it. |
LEAFY may therefore act as a “pioneer factor” capable of binding to the dense chromatin structure of certain genomic regions and initiating epigenetic changes that lead to gene expression. Indeed, some genes required to build floral organs such as petals or stamen are known to be under epigenetic repression and it makes sense that pioneer transcription factors would be required to initiate their expression. Other organisms contain transcription factors with similar oligomerisation domains. Our findings suggest that they might also act as pioneer transcription factors thanks to their oligomerisation domains. This work thus opens new avenues for understanding the role of these factors in the regulation of gene expression.
Principal publication and authors
A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor, C. Sayou (a), M.H. Nanao (b), M. Jamin (c), D. Posé (d,e), E. Thévenon (a), L. Grégoire (a), G. Tichtinsky (a), G. Denay (a), F. Ott (d), M. Peirats Llobet (a), M. Schmid (d,f), R. Dumas (a) and F. Parcy (a,g), Nature Communications 7, 11222 (2016); doi: 10.1038/ncomms11222.
(a) Laboratoire de Physiologie Cellulaire et Végétale, Université Grenoble Alpes, CNRS UMR5168, CEA/DRF/BIG, INRA UMR 1417, Grenoble (France)
(b) European Molecular Biology Laboratory, Grenoble (France)
(c) Institut de Biologie Structurale CEA/DRF, CNRS, Universite Grenoble Alpes, Grenoble (France)
(d) Department of Molecular Biology, Max Planck Institute for Developmental Biology, Tubingen (Germany)
(e) Instituto de Hortofruticultura Subtropical y Mediterranea, Universidad de Malaga (Spain)
(f) Umeå Plant Science Centre, Department of Plant Physiology, Umeå University (Sweden)
(g) Centre for Molecular Medicine and Therapeutics, Child and Family Research Institute, University of British Columbia, Vancouver (Canada)
References
[1] E. Moyroud et al., Trends in plant science 15, 346–52 (2010).
[2] C. Hamès et al., The EMBO journal 27, 2628–37 (2008).
[3] C. Kim and J.U. Bowie, Trends in biochemical sciences 28, 625–8 (2003).



