- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2017
- Structural biology
- Making rod-shaped bacteria
Making rod-shaped bacteria
The bacterial cell wall is a major antibiotic development target, and many pathogens require the 'elongasome' complex to guarantee its formation. MreC and PBPs form the core of the elongasome. Here, for the first time, a PBP:MreC complex has been characterised in atomic detail, revealing interaction regions that could be targeted by tailored inhibitors.
The peptidoglycan (PG) is an essential component of the bacterial cell wall, and plays a key role in shape maintenance, resistance to osmotic pressure, and cell division. Due to the central role PG plays in bacterial survival, its biosynthetic machinery has been a preferential target for antibiotic development for decades [1]. Proteins that are involved in PG biosynthesis associate in discrete multi-membered complexes that regulate cell division and elongation, and their inhibition or deregulation can lead to defects in cell shape and often cell wall lysis and death [2].
Penicillin-binding proteins (PBPs) catalyse the two last reactions in PG biosynthesis, and have been reported to interact with several members of the cell division and elongation complexes during the bacterial cell cycle [3]. One of these partners is MreC, a membrane-associated protein that has been shown to form fibres in certain bacteria and is thought to serve as a scaffold for macromolecules involved in cell wall elongation. However, protein interactions within these complexes have been reported to only form at defined points in the cell cycle, thus being fleeting in nature, fragile, and difficult to isolate.
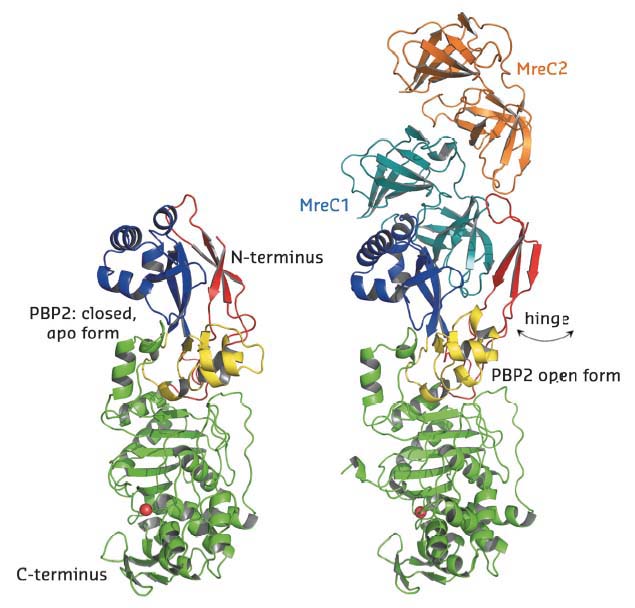
In this work, these difficulties were overcome and the PBP2:MreC core elongation complex from the human pathogen Helicobacter pylori was structurally and functionally characterised, as was PBP2 in its unbound form. All diffraction data were collected at beamlines BM30A and ID23-2. In the structure of PBP2 in its unbound form, its N-terminal region (blue and red in Figure 18) is ‘closed’, mimicking two clasped hands. In the structure of the PBP2:MreC complex, however, this region has undergone a conformational modification that has allowed it to open by approximately 80 Å, permitting binding of the partner molecule MreC (orange in Figure 18). Opening of this ‘hinge’ region reveals a non-polar platform that, when associated to MreC, forms a hydrophobic zipper.
 |
|
Fig. 18: Architecture and structural arrangements of PBP2 (left) and the PBP2:MreC complex (right) from Helicobacter pylori. In the structure of the complex, the N-terminus of PBP2 opens via a hinge region, exposing a previously hidden hydrophobic region that allows binding of MreC. The PBP2 active site, serine, is indicated as a red sphere. |
The importance of the stability of the zipper was verified by introducing mutations in vitro and in a microbiological setting. MreC forms that carried mutations in the hydrophobic zipper did not bind to PBP2, as verified by gel filtration and ITC. In addition, H. pylori cells that were modified to express the same mutations in their genome lost their characteristic elongated shape and rapidly increased in diameter, indicating that the PBP2:MreC hydrophobic zipper is essential for shape maintenance in bacteria.
This work thus provides the first visualisation of the core region of a bacterial elongation complex and reveals a novel area that could be targeted for the development of antibiotics that act by inhibiting cell wall elongation. This could lead to a generation of totally new molecules that are structurally distinct from antibiotics currently used in clinics and which face severe problems of resistance.
Principal publication and authors
Molecular architecture of the PBP2:MreC core bacterial cell wall synthesis complex, C. Contreras-Martel (a), A. Martins (b), C. Ecobichon (b), D. Maragno Trindade (c), P-J. Matteï (a), S. Hicham (b), P. Hardouin (a), M. El Ghachi (b), I. G. Boneca (b) and A. Dessen (a, c), Nat. Commun. 8, 776 (2017); doi: 10.1038/s41467-017-00783-2.
(a) Institut de Biologie Structurale (IBS), CNRS, CEA, Univ. Grenoble (France)
(b) Institut Pasteur & INSERM, Paris (France)
(c) Brazilian Biosciences National Laboratory (LNBio), CNPEM, Campinas, São Paulo (Brazil)
References
[1] L. L. Silver, Ann N. Y. Acad. Sci. 1277, 29-53 (2013).
[2] T. den Blaauwen et al., FEMS Microbiol. Rev. 32, 321-344 (2008).
[3] P.-J. Matteï et al., Curr. Opin. Struct. Biol. 20, 749-755 (2010).



